Response variable, specified as
a vector of length n, where n is the number of
observations. Each element of y is the response for the
corresponding row of X.
Missing values (NaN's) and infinite values (Inf's) are
allowed, but observations (rows) with these values will
automatically be excluded from the computations.
Data Types: single| double
Matrix of explanatory
variables (also called 'regressors') of dimension n x (p-1)
where p denotes the number of explanatory variables
including the intercept.
Rows of X represent observations, and columns represent
variables. By default, there is a constant term in the
model, unless it is explicitly removed using input option
intercept; so, do not include a column of 1s in X. Missing
values (NaN's) and infinite values (Inf's) are allowed,
since observations (rows) with such values will
automatically be excluded from the computations.
Data Types: single| double
Specify optional comma-separated pairs of Name,Value arguments.
Name is the argument name and Value
is the corresponding value. Name must appear
inside single quotes (' ').
You can specify several name and value pair arguments in any order as
Name1,Value1,...,NameN,ValueN.
Example:
'intercept',false
, 'family','YJ'
, 'nocheck',true
, 'conflev',0.99
, 'laseq',[-1:0.001;0.7]
, 'laseqPos',[-1:0.001;0.7]
, 'laseqNeg',[-1:0.001;0.7]
, 'plots',true
, 'usefmin',true
Indicator for the constant term (intercept) in the fit,
specified as comma-separated pair consisting of
'Intercept' and either true or false, to respectively
include or remove the constant term from the model.
Example: 'intercept',false
Data Types: boolean
String which
identifies the family of transformations which must be
used. Character. Possible values are 'BoxCox' (default),
'YJ' (Yao and Yohnson) and 'YJpn' (extended Yeo and
Johnson).
The Box-Cox family of power transformations equals
(y^{\lambda}-1)/\lambda for \lambda not equal to zero,
and \log(y) if \lambda = 0.
The Yeo-Johnson (YJ) transformation is the Box-Cox
transformation of y+1 for nonnegative values, and of
|y|+1 with parameter 2-lambda for y negative.
The extended Yeo-Johnson (YJpn) transformation is like
Yeo-Johnson, but admits two values of the transformation
parameters, respectively for positive and negative
observations.
Remark. BoxCox family can be used only if input y is
positive. Yeo-Johnson (and extended Yeo-Johnson family of
transformations do not have this limitation).
Example: 'family','YJ'
Data Types: char
If nocheck is equal to true no check is performed on vector y and matrix X. This
means that y and X are left unchanged. Note also that the
additional column of ones for the intercept is not added.
As default nocheck=false.
Example: 'nocheck',true
Data Types: boolean
The scalar is
between 0 and 1 and determines the confidence level for
lambda, based on the asymptotic chi1^2 of twice the
loglikelihood ratio. The default conflev value is 0.95;
Example: 'conflev',0.99
Data Types: double
Vector
which contains the sequence of values of lambda for
which the profile loglikelihood has to be computed if
family is 'BoxCox' or 'YJ'. The default value of laseq
is -2:0.001:2. This optional input is ignored if family
is 'YJpn';
Example: 'laseq',[-1:0.001;0.7]
Data Types: double
Vector which contains the sequence of values of
lambda which are used to transform positive observations
when family 'YJpn'. The default value of laseqPos is
-2:0.01:2. This optional input parameter is ignored if
family is 'BoxCox' or 'YJ';
Example: 'laseqPos',[-1:0.001;0.7]
Data Types: double
Vector which contains the sequence of values of
lambda which are used to transform negative observations
when family 'YJpn'. The default value of laseqNeg is
-2:0.01:2. This optional input is ignored if family is
'BoxCox' or 'YJ';
Example: 'laseqNeg',[-1:0.001;0.7]
Data Types: double
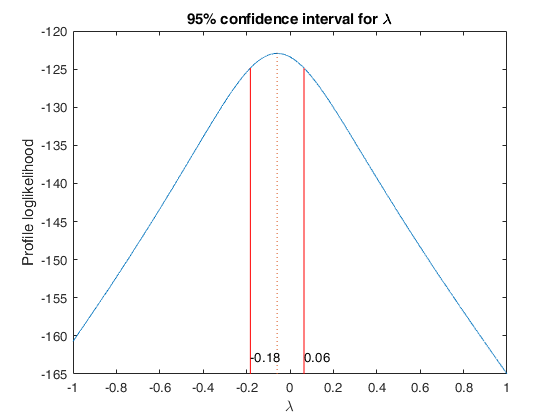
It specifies whether to show the profile log likelihood of
lambda. If plots is true, the plot of the profile
loglikelihood is produced together with the requested
confidence interval. The default value of prolik is false,
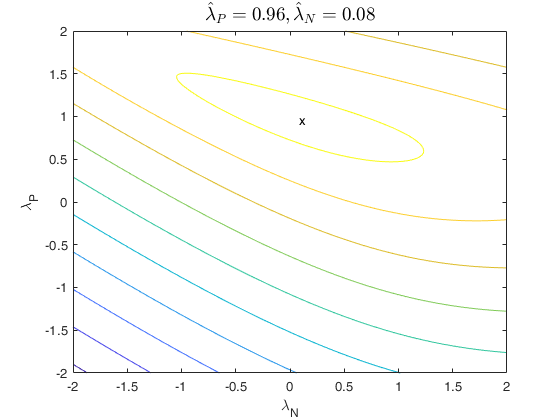
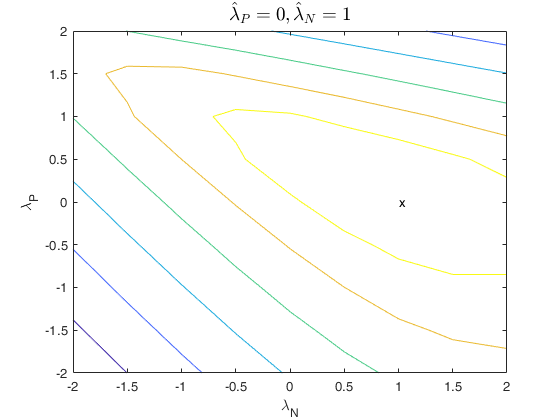
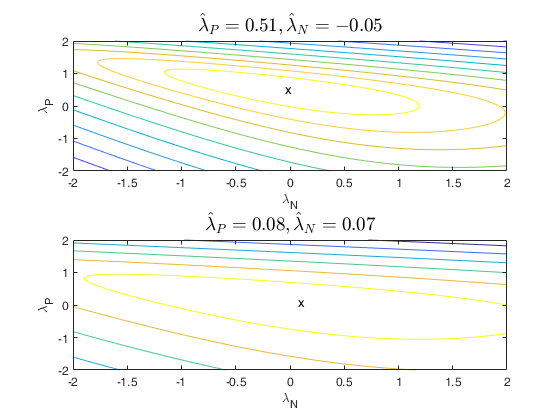
that is no plot is produced. If family is 'YJpn', a contour
plot is produced.
Example: 'plots',true
Data Types: boolean
This option applies only if family is YJpn. If usefmin is
true or usefmin is a struct, the maximum likelihood
estimates of \lambda_P and \lambda_N is computed using
the MATLAB solvers fminsearch or fminunc. The default value
of usefmin is false, that is the likelihood is evaluated at
the points laseqPos and laseqNeg without the solver.
If usefmin is a structure it may contain the following
fields:
| Value |
Description |
MaxIter |
Maximum number of iterations (default is 1000).
|
TolX |
Termination tolerance for the parameters
(default is 1e-7).
|
solver |
name of the solver. Possible values are
'fminsearch' (default) and 'fminunc'. fminunc needs the
optimization toolbox.
|
displayLevel |
amount of information displayed by
the algorithm. possible values are 'off' (displays no
information, this is the default), 'final' (displays
just the final output) and 'iter' (displays iterative
output to the command window).
|
Example: 'usefmin',true
Data Types: boolean or struct
 boxcoxR with all default options.
boxcoxR with all default options.