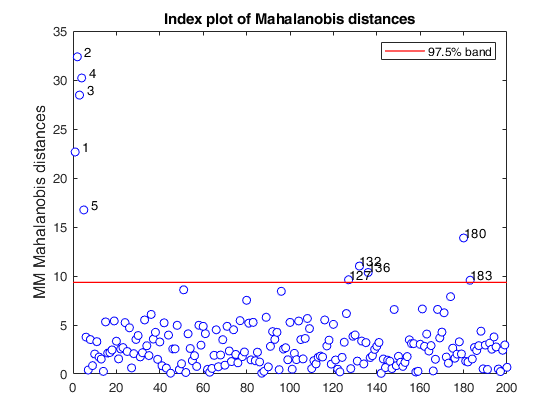
MMmultcore
MMmultcore computes multivariate MM estimators for a selected fixed scale
Syntax
Description
Examples
Input Arguments
Output Arguments
More About
References
Maronna, R.A., Martin D. and Yohai V.J. (2006), "Robust Statistics, Theory and Methods", Wiley, New York.
Acknowledgements
This function follows the lines of MATLAB/R code developed during the years by many authors. For more details see the R library robustbase http://robustbase.r-forge.r-project.org/.
The core of these routines, e.g. the resampling approach, however, has been completely redesigned, with considerable increase of the computational performance.
 MMmultcore with optional arguments.
MMmultcore with optional arguments.