mveeda
mveeda monitors Minimum volume ellipsoid for a series of values of bdp
Syntax
Description
Examples
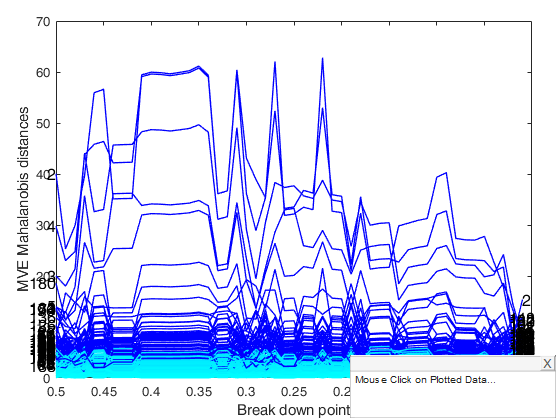
mveeda with all default options.
n=200;
v=3;
randn('state', 123456);
Y=randn(n,v);
% Contaminated data
Ycont=Y;
Ycont(1:5,:)=Ycont(1:5,:)+5;
RAW=mveeda(Ycont);
mveeda with optional arguments.
n=200;
v=3;
randn('state', 123456);
Y=randn(n,v);
% Contaminated data
Ycont=Y;
Ycont(1:5,:)=Ycont(1:5,:)+5;
RAW=mveeda(Ycont,'plots',1,'msg',0);
mve monitoring the reweighted estimates.
n=200;
v=3;
randn('state', 123456);
Y=randn(n,v);
% Contaminated data
Ycont=Y;
Ycont(1:5,:)=Ycont(1:5,:)+3;
[RAW,REW]=mveeda(Ycont);
Input Arguments
Y — Input data.
Matrix.
n x v data matrix; n observations and v variables. Rows of Y represent observations, and columns represent variables.
Missing values (NaN's) and infinite values (Inf's) are allowed, since observations (rows) with missing or infinite values will automatically be excluded from the computations.
Data Types: single|double
Name-Value Pair Arguments
Specify optional comma-separated pairs of Name,Value arguments.
Name is the argument name and Value
is the corresponding value. Name must appear
inside single quotes (' ').
You can specify several name and value pair arguments in any order as
Name1,Value1,...,NameN,ValueN.
'bdp',[0.5 0.4 0.3 0.2 0.1]
, 'nsamp',10000
, 'refsteps',0
, 'reftol',1e-8
, 'conflev',0.99
, 'nocheck',1
, 'plots',1
, 'msg',false
, 'ysaveRAW',1
, 'ysaveREW',1
bdp
—breakdown point.scalar | vector.
It measures the fraction of outliers the algorithm should resist. In this case any value greater than 0 but smaller or equal than 0.5 will do fine.
The default value of bdp is a sequence from 0.5 to 0.01 with step 0.01
Example: 'bdp',[0.5 0.4 0.3 0.2 0.1]
Data Types: double
nsamp
—Number of subsamples.scalar.
Number of subsamples of size v which have to be extracted (if not given, default = 500).
Example: 'nsamp',10000
Data Types: double
refsteps
—Number of refining iterations.scalar.
Number of refining iterationsin each subsample (default = 3).
refsteps = 0 means "raw-subsampling" without iterations.
Example: 'refsteps',0
Data Types: single | double
reftol
—scalar.default value of tolerance for the refining steps.
The default value is 1e-6;
Example: 'reftol',1e-8
Data Types: single | double
conflev
—Confidence level.scalar.
Number between 0 and 1 containing confidence level which is used to declare units as outliers.
Usually conflev=0.95, 0.975 0.99 (individual alpha) or 1-0.05/n, 1-0.025/n, 1-0.01/n (simultaneous alpha).
Default value is 0.975
Example: 'conflev',0.99
Data Types: double
nocheck
—Scalar.if nocheck is equal to 1 no check is performed on matrix Y.
As default nocheck=0.
Example: 'nocheck',1
Data Types: double
plots
—Plot on the screen.scalar.
If plots is equal to 1, it generates a plot which monitors raw mve Mahalanobis distances against values of bdp.
Example: 'plots',1
Data Types: double or structure
msg
—boolean.display | not messages on the screen.
If msg==true (default) messages are displayed on the screen about estimated time to compute the final estimator else no message is displayed on the screen.
Example: 'msg',false
Data Types: logical
ysaveRAW
—scalar that is set to 1 to request that the data matrix Y
is saved into the output structure RAW.this feature is meant at simplifying the use of function malindexplot.
Default is 0, i.e. no saving is done.
Example: 'ysaveRAW',1
Data Types: double
ysaveREW
—scalar that is set to 1 to request that the data matrix Y
is saved into the output structure REW.this feature is meant at simplifying the use of function malindexplot.
Default is 0, i.e. no saving is done.
Example: 'ysaveREW',1
Data Types: double
Output Arguments
RAW — description
Structure
Structure which contains the following fields
| Value | Description |
|---|---|
Loc |
length(bdp)-by-v matrix containing estimate of location for each value of bdp |
Cov |
v-by-v-by-length(bdp) 3D array containing robust estimate of covariance matrix for each value of bdp |
BS |
(v+1)-by-length(bdp) matrix containing the units forming best subset for each value of bdp |
MAL |
n x length(bdp) matrix containing the estimates of the robust Mahalanobis distances (in squared units) for each value of bdp |
Outliers |
n x length(bdp) matrix. Boolean matrix containing the list of the units declared as outliers for each value of bdp using confidence level specified in input scalar conflev |
Singsub |
Number of subsets without full rank. Notice that out.singsub > 0.1*(number of subsamples) produces a warning. |
Weights |
n x 1 vector containing the estimates of the weights. These weights determine which are the h observations which have been used to compute the final MVE estimates. |
bdp |
vector which contains the values of bdp which have been used |
h |
vector. Number of observations which have determined MVE for each value of bdp. |
Y |
Data matrix Y. |
class |
'mveeda'. This is the string which identifies the class of the estimator |
REW — description
Structure
Structure which contains the following fields:
| Value | Description |
|---|---|
Loc |
The robust location of the data, obtained after reweighting, if the RAW.cov is not singular. Otherwise the raw MVE center is given here. |
Cov |
The sequence of robust covariance matrices, obtained after reweighting and multiplying with a finite sample correction factor and an asymptotic consistency factor, if the raw MVE is not singular. |
MAL |
n x length(bdp) matrix containing the estimates of the robust Mahalanobis distances (in squared units) for each value of bdp |
Weights |
n x length(bdp) matrix containing the estimates of the weights. These weights determine which are the h observations which have been used to compute the final MVE estimates. |
Outliers |
A vector containing the list of the units declared as outliers after reweighting. |
Y |
Data matrix Y. |
class |
'mveeda' |
varargout —Indices
of the subsamples extracted for
computing the estimate.
C : matrix of size nsamp-by-v
More About
Additional Details
For each subset of v+1 observations \mu_J and C_J are the centroid and the covariance matrix based on subset J.
Rousseeuw and Leroy (RL) (eq. 1.25 chapter 7, p. 259) write the objective function for subset J as
RL_J=\left( med_{i=1, ..., n} \sqrt{ (y_i -\mu_J)' C_J^{-1} (y_i -\mu_J) } \right)^v \sqrt|C_J| Maronna Martin and Yohai (MMY), eq. (6.57), define \Sigma_J = C_j / |C_j|^{1/v} and write the objective function for subset J as MMY_J = \hat \sigma \left( (y_i -\mu_J)' \Sigma_J^{-1} (y_i -\mu_J) \right) |C_J|^{1/v} = \hat \sigma \left( (y_i -\mu_J)' C_J^{-1} (y_i -\mu_J) \right) |C_J|^{1/v}where \hat \sigma \left( (y_i -\mu_J)' C_J^{-1} (y_i -\mu_J) \right) = med_{i=1, ..., n}(y_i -\mu_J)' C_J^{-1} (y_i -\mu_J).
Note that MMY_J= (RL)^{2/v}.
To RAW.cov a consistency factor has been applied which is based on chi2inv(1-bdp,v). On the other hand to REW.cov the usual asymptotic consistency factor is applied. In this case we have used the empirical percentage of trimming that is the ratio hemp/n where hemp is the number of units which had a MD smaller than the cutoff level determined by thresh=chi2inv(conflev,v); MD are computed using RAW.loc and RAW.cov.
The mve method is intended for continuous variables, and assumes that the number of observations n is at least 5 times the number of variables v.
References
Rousseeuw, P.J. (1984), Least Median of Squares Regression, "Journal of the American Statistical Association", Vol. 79, pp. 871-881.
Rousseeuw, P.J. and Leroy A.M. (1987), Robust regression and outlier detection, Wiley New York.
Acknowledgements
This function follows the lines of MATLAB/R code developed during the years by many authors.
For more details see the R library robustbase http://robustbase.r-forge.r-project.org/ The core of these routines, e.g. the resampling approach, however, has been completely redesigned, with considerable increase of the computational performance.
 mve monitoring with varargout.
mve monitoring with varargout.