wthin
wthin thins a uni/bi-dimensional dataset
Syntax
Description
Computes retention probabilities and bernoulli (0/1) weights on the basis of data density estimate.
Bi-dimensional thinning.Wt
=wthin(X,
Name, Value)
Examples
Univariate thinning.
clear all; close all;
% The dataset is bi-dimensional and contain two collinear groups with
% regression structure. One group is dense, with 1000 units; the second
% has 100 units. Thinning in done according to the density of the values
% predicted by the OLS fit.
x1 = randn(1000,1);
x2 = 8 + randn(100,1);
x = [x1 ; x2];
y = 5*x + 0.9*randn(1100,1);
b = [ones(1100,1) , x] \ y;
yhat = [ones(1100,1) , x] * b;
plot(x,y,'.',x,yhat);
%x3 = 0.2 + 0.01*randn(1000,1);
%y3 = 40 + 0.01*randn(1000,1);
%plot(x,y,'.',x,yhat,'--',x3,y3,'.');
% thinning over the predicted values
%[Wt,pretain] = wthin([yhat ; y3], 'retainby','comp2one');
% thinning over the predicted values when specifying a thinning
%probability pstar (randomized thinning).
pstar=0.95
[Wt,pretain] = wthin(yhat, 'retainby','comp2one','pstar',pstar);
% thinning over the predicted values when specifying a thinning
%cup (winsorized thinning).
cup=0.5
[Wt,pretain] = wthin(yhat, 'retainby','comp2one','cup',cup);
figure;
plot(x(Wt,:),y(Wt,:),'k.',x(~Wt,:),y(~Wt,:),'r.');
drawnow;
axis manual;
title('univariate thinning over predicted ols values')
clickableMultiLegend(['Retained: ' num2str(sum(Wt))],['Thinned: ' num2str(sum(~Wt))]);
Bi-dimensional thinning.
Same dataset, but thinning is done on the original bi-variate data.
x1 = randn(1000,1);
x2 = 8 + randn(100,1);
x = [x1 ; x2];
y = 5*x + 0.9*randn(1100,1);
b = [ones(1100,1) , x] \ y;
plot(x,y,'.');
% thinning over the original bi-variate data
[Wt2,pretain2] = wthin([x,y]);
plot(x(Wt2,:),y(Wt2,:),'k.',x(~Wt2,:),y(~Wt2,:),'r.');
drawnow;
axis manual;
title('bivariate thinning')
clickableMultiLegend(['Retained: ' num2str(sum(Wt2))],['Thinned: ' num2str(sum(~Wt2))]);
Use of 'retainby' option.
Since the thinning on the original bi-variate data with the default retention method ('inverse') removes too many units, let's try with the less conservative 'comp2one' option.
x1 = randn(1000,1);
x2 = 8 + randn(100,1);
x = [x1 ; x2];
y = 5*x + 0.9*randn(1100,1);
b = [ones(1100,1) , x] \ y;
plot(x,y,'.');
% thinning over the original bi-variate data
[Wt2,pretain2] = wthin([x,y], 'retainby','comp2one');
plot(x(Wt2,:),y(Wt2,:),'k.',x(~Wt2,:),y(~Wt2,:),'r.');
drawnow;
axis manual
clickableMultiLegend(['Retained: ' num2str(sum(Wt2))],['Thinned: ' num2str(sum(~Wt2))]);
title('"comp2one" thinning over the original bi-variate data');
Optional output Xt.
Same dataset, the retained data are also returned using varagout option.
x1 = randn(1000,1);
x2 = 8 + randn(100,1);
x = [x1 ; x2];
y = 5*x + 0.9*randn(1100,1);
% thinning over the original bi-variate data
[Wt2,pretain2,RetUnits] = wthin([x,y]);
% disp(RetUnits)
Related Examples
thinning on the fishery dataset.
load fishery;
X=fishery{:,:};
% some jittering is necessary because duplicated units are not treated
% in tclustreg: this needs to be addressed
X = X + 10^(-8) * abs(randn(677,2));
% thinning over the original bi-variate data
[Wt3,pretain3,RetUnits3] = wthin(X ,'retainby','comp2one');
figure;
plot(X(Wt3,1),X(Wt3,2),'k.',X(~Wt3,1),X(~Wt3,2),'rx');
drawnow;
axis manual
clickableMultiLegend(['Retained: ' num2str(sum(Wt3))],['Thinned: ' num2str(sum(~Wt3))]);
title('"comp2one" thinning on the fishery dataset');
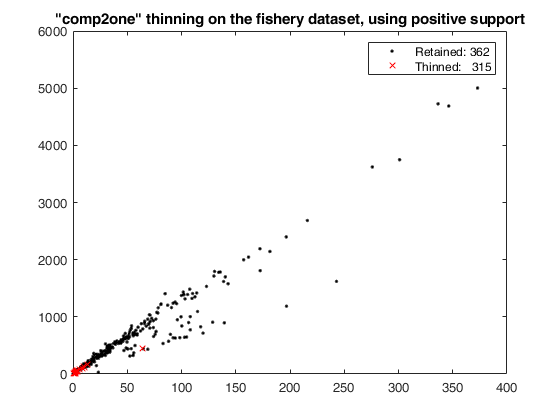
 thinning on the fishery dataset using 'positive' support.
thinning on the fishery dataset using 'positive' support.
 thinning on the fishery dataset using 'positive' support.
thinning on the fishery dataset using 'positive' support.
load fishery;
X=fishery{:,:};
% some jittering is necessary because duplicated units are not treated
% in tclustreg: this needs to be addressed
X = X + 10^(-8) * abs(randn(677,2));
% thinning over the original bi-variate data
[Wt3,pretain3,RetUnits3] = wthin(X ,'retainby','comp2one','support','positive');
figure;
plot(X(Wt3,1),X(Wt3,2),'k.',X(~Wt3,1),X(~Wt3,2),'rx');
drawnow;
axis manual
clickableMultiLegend(['Retained: ' num2str(sum(Wt3))],['Thinned: ' num2str(sum(~Wt3))]);
title('"comp2one" thinning on the fishery dataset, using positive support');
univariate thinning with less than 100 units.
As the first examp[le above, but with less than 100 units in the data.
x1 = randn(850,1);
x2 = 8 + randn(10,1);
x = [x1 ; x2];
y = 5*x + 0.9*randn(860,1);
b = [ones(860,1) , x] \ y;
yhat = [ones(860,1) , x] * b;
plot(x,y,'.',x,yhat,'--');
% thinning over the predicted values
[Wt,pretain] = wthin(yhat, 'retainby','comp2one');
plot(x(Wt,:),y(Wt,:),'k.',x(~Wt,:),y(~Wt,:),'r.');
drawnow;
axis manual
title('univariate thinning over ols values predicted on a small dataset')
clickableMultiLegend(['Retained: ' num2str(sum(Wt))],['Thinned: ' num2str(sum(~Wt))]);
Input Arguments
X — Input data.
Vector or 2-column matrix.
The structure contains the uni/bi-variate data to be thinned on the basis of a probability density estimate.
Data Types: single| double
Name-Value Pair Arguments
Specify optional comma-separated pairs of Name,Value arguments.
Name is the argument name and Value
is the corresponding value. Name must appear
inside single quotes (' ').
You can specify several name and value pair arguments in any order as
Name1,Value1,...,NameN,ValueN.
bandwidth,0.35
, support,'positive'
, cup, 0.8
, pstar, 0.95
, 'method','comp2one'
bandwidth
—bandwidth value.scalar.
The bandwidth used to estimate the density. It can be estimated from the data using function bwe.
Example: bandwidth,0.35
Data Types: scalar
support
—support value.character array.
The support of the density estimation step. It can be 'unbounded' (the default) or 'positive' if the data are left-truncated with long right tails. In the latter case, the option performs the density estimate in the log domain and then transform the result back. The theoretical rationale is that when kernel density is applied to positive data, it does not yield proper PDFs.
Example: support,'positive'
Data Types: char
cup
—pdf upper limit.scalar.
The upper limit for the pdf used to compute the retantion probability. If cup = 1 (default), no upper limit is set.
Example: cup, 0.8
Data Types: scalar
pstar
—thinning probability.scalar.
Probability with each a unit enters in the thinning procedure. If pstar = 1 (default), all units enter in the thinning procedure.
Example: pstar, 0.95
Data Types: scalar
retainby
—retention method.string.
The function used to retain the observations. It can be: - 'inverse' , i.e. (1 ./ pdfe) / max((1 ./ pdfe))) - 'comp2one' (default), i.e. 1 - pdfe/max(pdfe))
Example: 'method','comp2one'
Data Types: char
Output Arguments
Wt —vector of Bernoulli weights.
Vector
Contains 1 for retained units and 0 for thinned units.
Data Types - single | double.
pretain —vector of retention probabilities.
Vector
These are the probabilities that each point in X will be retained, estimated using a gaussian kernel using function ksdensity.
Data Types - single | double.
References
Bowman, A.W. and Azzalini, A. (1997), "Applied Smoothing Techniques for Data Analysis", Oxford University Press.
Wand, M.P. and Marron, J.S. and Ruppert, D. (1991), "Transformations in density estimation", Journal of the American Statistical Association, 86(414), 343-353.
See Also
ksdensity
|
mvksdensity
|
bwe