Desired number of variables.
Data Types: int16|int32|int64|single|double
Desired number of groups.
Data Types: int16|int32|int64|single|double
Structure containing the following fields:
| Value |
Description |
pars |
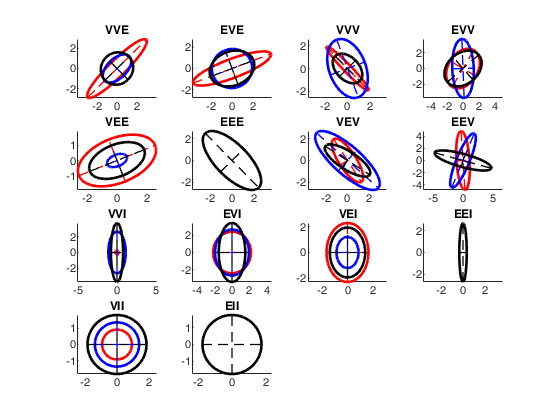
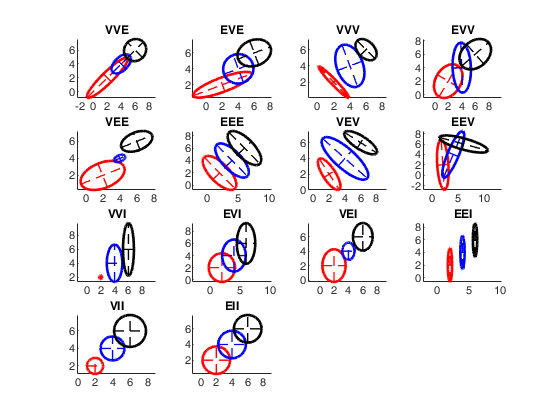
type of Gaussian Parsimonious Clustering Model. Character.
A 3 letter word in the set:
'VVE','EVE','VVV','EVV','VEE','EEE','VEV','EEV','VVI',
'EVI','VEI','EEI','VII','EII'.
The field pa.pars is compulsory. All the other fields are
non necessary. If they are not present they are set to
their default values.
|
exactrestriction |
boolean. If pa.exactrestriction is true
the covariance matrices have to be generated with the exact
values of the restrictions specified in pa.cdet, pa.shw and
pa.swb. In order to reach this purpose, this procedure
ensures to generate shape matrices in which at least one
ratio of the elements inside each component is greater or
equal than that specified is pa.shw and at least one ratio
among the ordered elements of each shape matrix is greater or
equal tahn pa.shb. The successive application of routine
restrSgimaGPCM guarrantes that the inequalites become
equalities. The default value of pa.exaxtrestriction is
false therefore covariance matrices are generated without
implying any constraint. If pa.exactrestriction is true
covariance matrices are generated with:
1) max ratio between determinants equal to pa.cdet (if
pa.cdet is specififed);
2) at least a ratio between the elements of each shape matrix greater or equal than
pa.shw (if pa.shw is specififed);
3) max ratio between the ordered elements of each shape
matrices greater or equal than pa.shb).
|
cdet |
scalar in the interval [1 Inf) which specifies the
the restriction which has to be applied to the determinants.
This field is used just if pa.exactrestriction is true.
|
shw |
scalar in the interval [1 Inf) which specifies the
the restriction which has to be applied to the elements of
the shape matrices inside each group. This field is used
just if pa.exactrestriction is true.
|
shb |
scalar in the interval [1 Inf) which specifies the
the restriction which has to be applied to the ordered elements of
the shape matrices across groups. This field is used
just if pa.exactrestriction is true.
|
Data Types: struct


 Covariance matrices contours for the 14 models.
Covariance matrices contours for the 14 models.