spmplot
spmplot produces an interactive scatterplot matrix with boxplots or histograms on the main diagonal and possibly robust bivariate contours
Syntax
Description
Call of spmplot with option plo: the TickLabels are formatted.H
=spmplot(Y,
Name, Value)
Examples
Call of spmplot with Y as table.
In this case the VariableNames of the table are added at the margins of the plot.
load fisheriris;
meastable=array2table(meas,'VariableNames',{'SL','SW','PL','PW'});
spmplot(meastable);
Call of spmplot with option plo: the TickLabels are formatted.
load fisheriris; % labels determined automatically spmplot(meas); % labels are formatted plo=struct; plo.TickLabels = '%.2f'; figure; spmplot(meas,'plo',plo); % labels are deleted plo=struct; plo.TickLabels = []; figure; spmplot(meas,'plo',plo);
Related Examples
Example of use of options 'nameY', 'nameYlength' and 'nameYrot'.
In this example these options are passed as Name/Values.
nameY=["aaaawew" "bbccccc" "ccdddd"]; % Truncated the name of the variables to 3 characters and rotate % the labels by 45 degrees. spmplot(randn(30,3),'nameY',nameY,'nameYlength',3,'nameYrot',45);
Call of spmplot without name/value pairs.
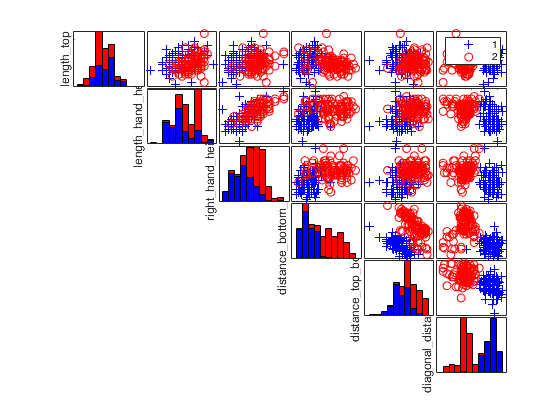
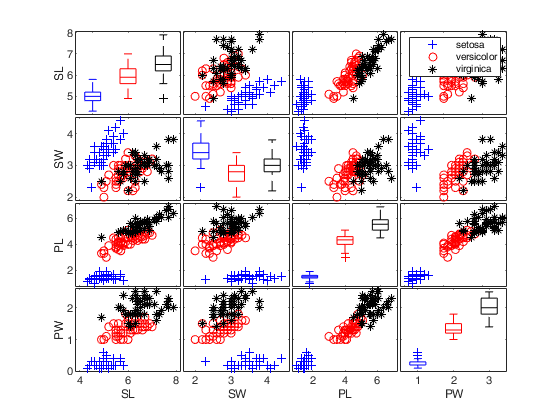
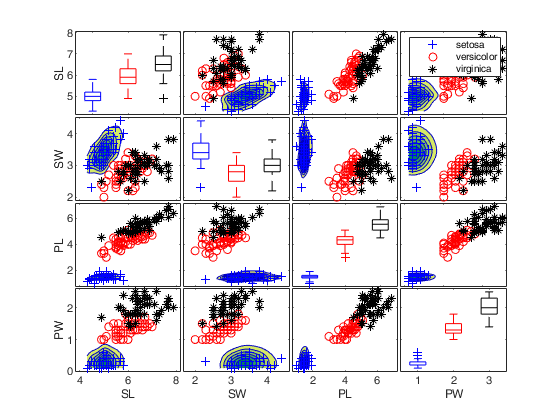
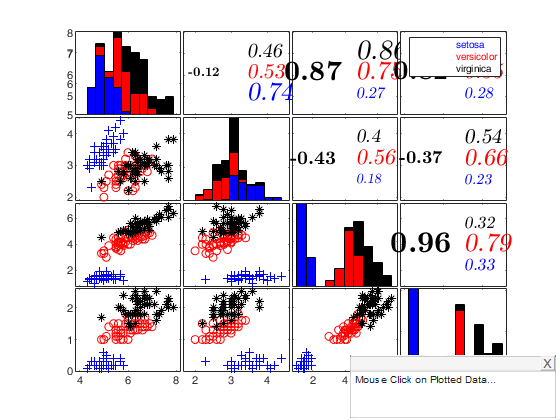
Iris data: scatter plot matrix with univariate boxplots on the main diagonal.
close all
load fisheriris;
plo=struct;
plo.nameY={'SL','SW','PL','PW'};
figure;
spmplot(meas,species,plo,'hist');
Call of spmplot without name/value pairs (2nd example).
With this way of calling spmplot just the first 4 arguments are considered. All the rest is discarded. A message appears to alert the user that this is the case.
close all
load fisheriris;
plo=struct;
plo.nameY={'SL','SW','PL','PW'};
figure;
spmplot(meas,species,plo,'hist','tag','dfgdfg');
 Call of spmplot with name/value pairs.
Call of spmplot with name/value pairs.
 Call of spmplot with name/value pairs.
Call of spmplot with name/value pairs.Specifying overlay, also discarding some groups with the field include, and changing the default colormap.
% The Tag setting will be used in the next example to demonstrate the
% undock option.
% Iris data: scatter plot matrix with univariate boxplots on the main
% diagonal.
close all
load fisheriris;
plo=struct;
plo.nameY={'SL','SW','PL','PW'};
spmplot(meas,'group',species,'plo',plo,'dispopt','box');
figure
spmplot(meas,'group',species,'plo',plo,'dispopt','box','overlay','ellipse');
figure
spmplot(meas,'group',species,'plo',plo,'dispopt','box','overlay','contour');
figure
spmplot(meas,'group',species,'plo',plo,'dispopt','box','overlay','contourf');
set(gcf,'Tag','newTag')
cascade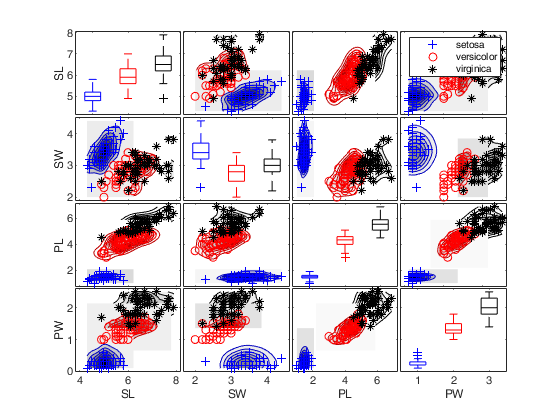
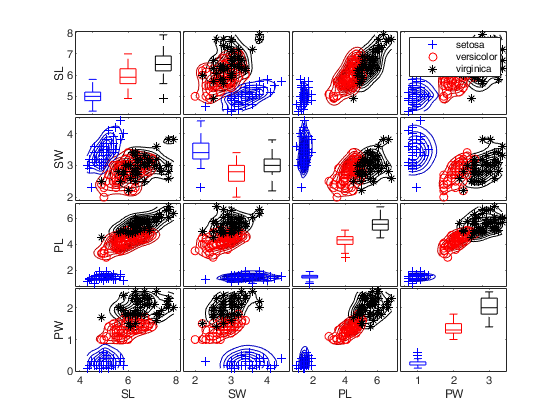
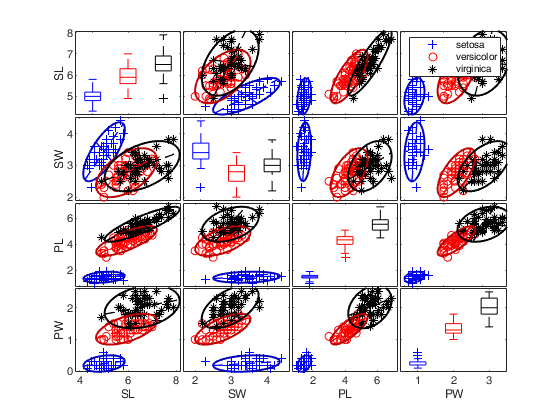
 Call of spmplot with name/value pairs and specifying overlay and undock.
Call of spmplot with name/value pairs and specifying overlay and undock.
 Call of spmplot with name/value pairs and specifying overlay and undock.
Call of spmplot with name/value pairs and specifying overlay and undock.The latter argument requires to change the tag of the scatterplot matrix not to delete.
% This example uses a matrix of logicals to set the undocked panels
load fisheriris;
plo=struct;
plo.nameY={'SL','SW','PL','PW'};
figure
spmplot(meas,'group',species,'plo',plo,'dispopt','hist','undock',logical(eye(size(meas,2))));
cascade
% This example uses a matrix n x 2 to set the undocked panels
close all;
figure
spmplot(meas,'group',species,'plo',plo,'dispopt','box','overlay','boxplotb','undock',[1,3;2,4]);
cascade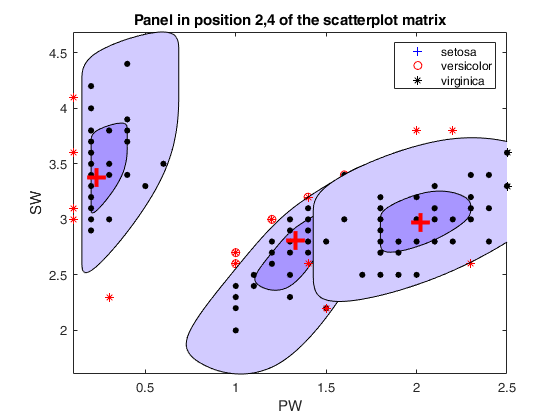
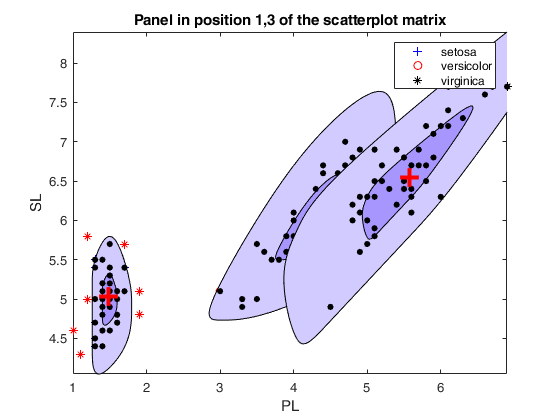
 Call of spmplot with name/value pairs and additional options for
overlay, specifying densities just for one group.
Call of spmplot with name/value pairs and additional options for
overlay, specifying densities just for one group.
 Call of spmplot with name/value pairs and additional options for
overlay, specifying densities just for one group.
Call of spmplot with name/value pairs and additional options for
overlay, specifying densities just for one group.
% Iris data: scatter plot matrix with univariate boxplots on the main
% diagonal.
close all
load fisheriris;
plo=struct;
plo.nameY={'SL','SW','PL','PW'};
over = struct;
over.type = 'contourf';
over.include = logical([1 0 0]);
over.cmap = summer;
figure
spmplot(meas,'group',species,'plo',plo,'dispopt','box','overlay',over);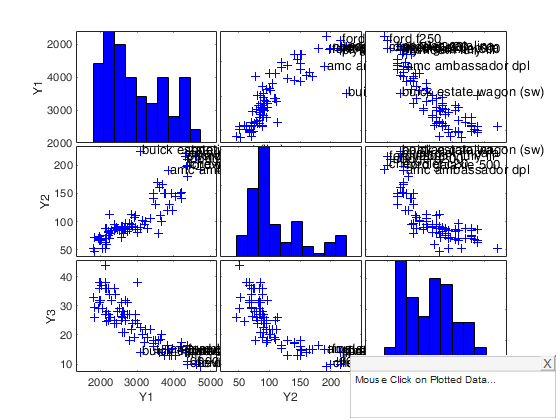
Iris data: scatter plot matrix with univariate boxplots on the main
diagonal and personalized options for symbols, colors, symbol
size and no legend.
close all;
load fisheriris;
plo=struct;
plo.nameY={'SL','SW','PL','PW'}; % Name of the variables
plo.clr='kbr'; % Colors of the groups
plo.sym={'+' '+' 'v'}; % Symbols of the groups (inside a cell)
% Symbols can also be specified as characters
% plo.sym='++v'; % Symbols of the groups
plo.siz=3.4; % Symbol size
plo.doleg='off'; % Remove the legend
figure
spmplot(meas,species,plo,'box');
Example of spmplot called by routine FSM.
Generate contaminated data.
close all;
state=100;
randn('state', state);
n=200;
Y=randn(n,3);
Ycont=Y;
Ycont(1:5,:)=Ycont(1:5,:)+3;
% spmplot is called automatically by all outlier detection methods, e.g. FSM
[out]=FSM(Ycont,'plots',1);
Now test the direct use of FSM.
Set two groups, e.g. those obtained from FSM.
% Generate contaminated data
state=100;
randn('state', state);
n=200;
Y=randn(n,3);
Ycont=Y;
Ycont(1:5,:)=Ycont(1:5,:)+3;
close all;
[out]=FSM(Ycont,'plots',1);
group = zeros(n,1);
group(out.outliers)=1;
plo=struct;
plo.labeladd='1'; % option plo.labeladd is used to label the outliers
% By default, the legend identifies the groups with the identifiers
% given in vector 'group'.
figure;
plo.clr = 'br';
spmplot(Ycont,group,plo,'box');
spm with personalized tags.
With two groups, and if the Tag of the figure contains the word 'outlier', the legend will identify one group for outliers and the other for normal units. The largest number in the 'group' variable identifies the group of outliers.
close all
state=100;
randn('state', state);
n=200;
Y=randn(n,3);
Ycont=Y;
Ycont(1:5,:)=Ycont(1:5,:)+3;
[out]=FSM(Ycont,'plots',1);
group = zeros(n,1);
group(out.outliers)=1;
plo=struct;
plo.labeladd='1'; % option plo.labeladd is used to label the outliers
figure('tag','This is a scatterplot with ouTliErs'); % case insensitive
spmplot(Ycont,group);
% If the Tag of the Figure contains the string 'group', then the
% legend identifies the groups with 'Group 1', Group 2', etc.
figure('tag','This scatterplot contains groups');
spmplot(Ycont,group,plo,'box');
% If the tag figure includes the word 'brush', the legend will identify
% one group for 'Unbrushed units' and the others for 'Brushed units 1',
% 'Brushed units 2', etc.
figure('Tag','Scatterplot with brushed units');
spmplot(Ycont,group,plo);
cascade;
An example with 5 groups.
close all
rng('default')
rng(2); n1=100;
n2=80;
n3=50;
n4=80;
n5=70;
v=5;
Y1=randn(n1,v)+5;
Y2=randn(n2,v)+3;
Y3=rand(n3,v)-2;
Y4=rand(n4,v)+2;
Y5=rand(n5,v);
group=ones(n1+n2+n3+n4+n5,1);
group(n1+1:n1+n2)=2;
group(n1+n2+1:n1+n2+n3)=3;
group(n1+n2+n3+1:n1+n2+n3+n4)=4;
group(n1+n2+n3+n4+1:n1+n2+n3+n4+n5)=5;
Y=[Y1;Y2;Y3;Y4;Y5];
spmplot(Y,group,[],'box');
Another example of spmplot called with name/pairs.
The example which follow make use of the name/value pairs arguments.
close all
load fisheriris;
plo=struct;
plo.nameY={'SL','SW','PL','PW'}; % Name of the variables
plo.clr='kbr'; % Colors of the groups
plo.sym={'+' '+' 'v'}; % Symbols of the groups (inside a cell)
% Symbols can also be specified as characters
% plo.sym='++v'; % Symbols of the groups
plo.siz=3.4; % Symbol size
spmplot(meas,'group',species,'plo',plo,'dispopt','box','tag','myspm');
Interactive example 1.
In the previous examples the first argument of spmplot was a matrix. In the two examples below the first argument is a structure which contains the fields Y and Un Example when first input argument is a structure.
% Example of use of option databrush
close all
rng(841,'shr3cong');
n=100;
v=3;
m0=v+1;
Y=randn(n,v);
% Contaminated data
Ycont=Y;
Ycont(1:5,:)=Ycont(1:5,:)+3;
[fre]=unibiv(Y);
%create an initial subset with the 3 observations with the lowest
%Mahalanobis Distance
fre=sortrows(fre,4);
bs=fre(1:m0,1);
[out]=FSMeda(Ycont,bs,'plots',1);
% mmdplot(out);
figure
% 'Label' 'on' 'RemoveLabels' 'off' enables the user to label the units in the scatterplot
% matrix once selected. Option labeladd '1' inside databrush enables to add
% the labels of the selected units in the linked malfwdplot
spmplot(out,'databrush',{'persist','on','selectionmode' 'Rect', ...
'Label' 'on' 'RemoveLabels' 'off' 'labeladd','1'},'dispopt','hist');
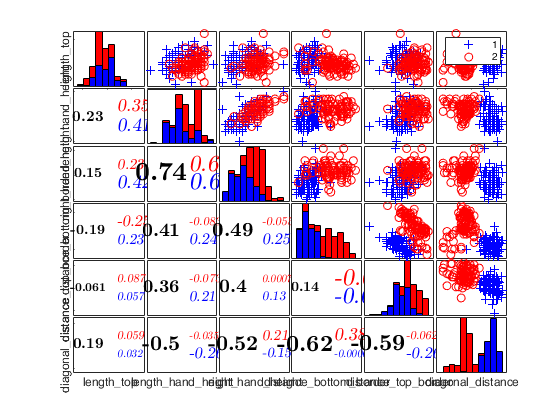
Example of use of option datatooltip.
First input argument is a structure.
close all n=100; v=3; m0=3; Y=randn(n,v); % Contaminated data Ycont=Y; Ycont(1:10,:)=5; [fre]=unibiv(Ycont); %create an initial subset with the 3 observations with the lowest %Mahalanobis Distance fre=sortrows(fre,4); bs=fre(1:m0,1); [out]=FSMeda(Ycont,bs,'plots',1); % mmdplot(out); figure plo=struct; plo.labeladd='1'; plo.clr = 'b'; spmplot(out,'datatooltip',1,'plo',plo);
First example of the use of options selunit when first input argument is a 2D array.
In this case row names of Y are not specified so numbers in the interval 1:n are added to the scatters.
n=100; p=5; seluni=[10 30]; Y =randn(n,p); Y(seluni,:)=Y(seluni,:)+2; % add labels for units inside vector seluni spmplot(Y,'selunit',seluni);
Second example of the use of options selunit when first input argument is a 2D array.
In this case the row names are contained inside input argument plo.label
close all load carsmall x1 = Weight; x2 = Horsepower; % Contains NaN data y = MPG; % response Y=[x1 x2 y]; % Remove Nans boo=~isnan(y); Y=Y(boo,:); RowLabelsMatrixY=Model(boo,:); seluni=[10 30]; plo=struct; plo.label=cellstr(RowLabelsMatrixY); % add labels for units inside vector seluni spmplot(Y,'selunit',seluni,'plo',plo);
 Option datatooltip combined with rownames.
Option datatooltip combined with rownames.
 Option datatooltip combined with rownames.
Option datatooltip combined with rownames.Example of use of option datatooltip.
% First input argument is a structure. close all load carsmall x1 = Weight; x2 = Horsepower; % Contains NaN data y = MPG; % response Y=[x1 x2 y]; % Remove Nans boo=~isnan(y); Y=Y(boo,:); Model=Model(boo,:); m0=5; [fre]=unibiv(Y); %create an initial subset with the 3 observations with the lowest %Mahalanobis Distance fre=sortrows(fre,4); bs=fre(1:m0,1); [out]=FSMeda(Y,bs,'plots',0); % field label (rownames) is added to structure out % In this case datatooltip will display the rowname and not the default % string row. out.label=cellstr(Model); figure plo=struct; plo.labeladd='1'; plo.clr = 'b'; spmplot(out,'datatooltip',1,'plo',plo); % The units which are already labelled in each panel of the scatter % plot matrix are those which in the search had a Mahalanobis distance % greater than 2.5. Note that the labelling is controlled by option selunit.
Interactive example 2.
Example of the use of options selstep and selunit combined with databrush.
% selunit is passed as a character.
% It produces a scatter plot matrix in which labels are put for units
% which have a Mahalanobis distance greater than str2num(selunit). When a set of
% units is brushed in the spmplot in the monitoring MD
% plot the labels for the units which have a MD greater than 10
% are added in steps selsteps.
load carsmall
x1 = Weight;
x2 = Horsepower; % Contains NaN data
y = MPG; % response
Y=[x1 x2 y];
% Remove Nans
boo=~isnan(y);
Y=Y(boo,:);
Model=Model(boo,:);
m0=5;
[fre]=unibiv(Y);
fre=sortrows(fre,4);
bs=fre(1:m0,1);
[out]=FSMeda(Y,bs,'plots',0);
spmplot(out,'selstep',[60 80],'selunit','10',...
'databrush',{'persist','off','selectionmode' 'Rect'});
Interactive example 3.
Example of the use of options selstep and selunit.
% selunit is passed as a numeric vector.
% It produces a scatter plot matrix in which labels are put for units
% selunit. When a set of
% units is brushed in the spmplot in the monitoring MD
% plot the labels for the units selunit
% are added in steps selsteps.
load carsmall
x1 = Weight;
x2 = Horsepower; % Contains NaN data
y = MPG; % response
Y=[x1 x2 y];
% Remove Nans
boo=~isnan(y);
Y=Y(boo,:);
Model=Model(boo,:);
m0=5;
[fre]=unibiv(Y);
fre=sortrows(fre,4);
bs=fre(1:m0,1);
[out]=FSMeda(Y,bs,'plots',0);
spmplot(out,'selstep',[60 80],'selunit',1:5,...
'databrush',{'persist','off','selectionmode' 'Rect'});
Interactive example 4.
Example of the use of options selstep and selunit.
% selunit is passed as a cell array of length 2.
% It produces a scatter plot matrix in which labels are put for units
% which have a min MD > selunit{1} and max MD <selunit{2}. When a set of
% units is brushed in the spmplot in the monitoring MD
% plot the labels for the units selunit
% are added in steps selsteps.
load carsmall
x1 = Weight;
x2 = Horsepower; % Contains NaN data
y = MPG; % response
Y=[x1 x2 y];
% Remove Nans
boo=~isnan(y);
Y=Y(boo,:);
Model=Model(boo,:);
m0=5;
[fre]=unibiv(Y);
fre=sortrows(fre,4);
bs=fre(1:m0,1);
[out]=FSMeda(Y,bs,'plots',0);
spmplot(out,'selstep',[60 80],'selunit',{'1.2' '1.6'},...
'databrush',{'persist','off','selectionmode' 'Rect'});
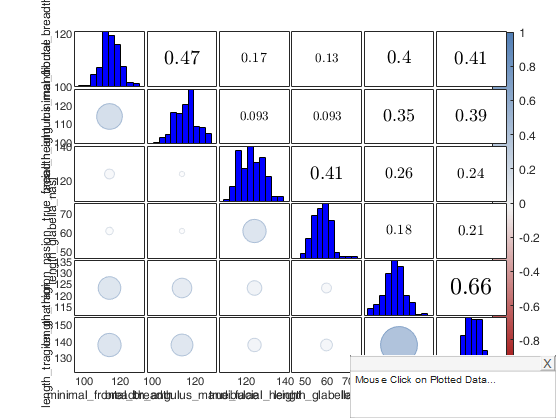
 Example 1 of use of option typespm passed as struct with groups.
Example 1 of use of option typespm passed as struct with groups.
 Example 1 of use of option typespm passed as struct with groups.
Example 1 of use of option typespm passed as struct with groups.
close all load swiss_banknotes.mat X=swiss_banknotes; group=ones(200,1); group(101:end)=2; plo = struct; plo.TickLabels = []; % In the lower part the correlations are shown with numbers typespm=struct; typespm.lower="number"; typespm.upper="scatter"; spmplot(swiss_banknotes,'plo',plo,'group',group,'typespm',typespm);
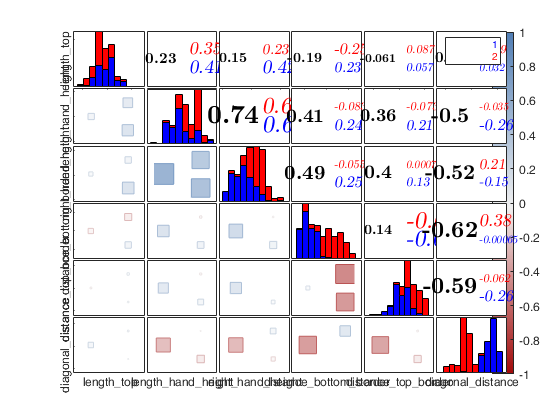
 Example 2 of use of option typespm passed as struct with groups.
Example 2 of use of option typespm passed as struct with groups.
 Example 2 of use of option typespm passed as struct with groups.
Example 2 of use of option typespm passed as struct with groups.
close all load swiss_banknotes.mat X=swiss_banknotes; group=ones(200,1); group(101:end)=2; plo = struct; plo.TickLabels = []; % In the lower part the correlations are shown with squares % and in the upper part with numbers typespm=struct; typespm.lower="square"; typespm.upper="number"; spmplot(swiss_banknotes,'group',group,'plo',plo,'typespm',typespm);
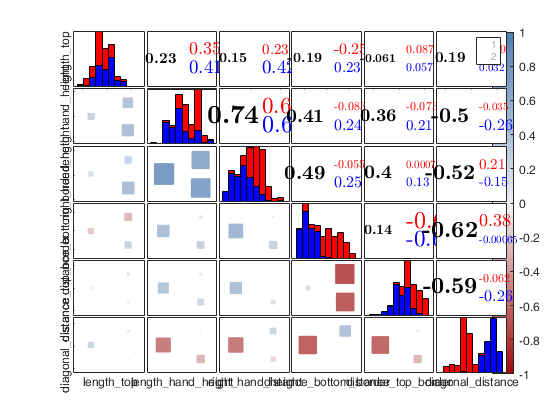
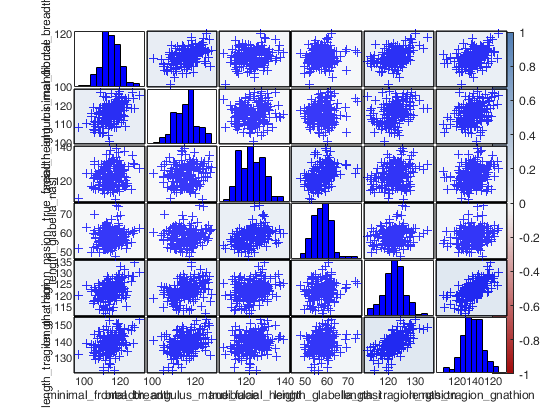
 Example 3 of use of option typespm with lower = "none".
Example 3 of use of option typespm with lower = "none".
 Example 3 of use of option typespm with lower = "none".
Example 3 of use of option typespm with lower = "none".
close all load swiss_banknotes.mat X=swiss_banknotes; group=ones(200,1); group(101:end)=2; plo = struct; plo.TickLabels = []; % In the lower part the correlations are shown with numbers typespm=struct; typespm.lower="none"; typespm.upper="scatter"; spmplot(swiss_banknotes,'group',group,'plo',plo,'typespm',typespm);
Input Arguments
Y — data matrix (2D array or table) containing n observations on v variables
or a structure 'out' coming from function FSMeda.
Matrix or
struct.
If Y is a 2D array or table, varargin can be either a sequence of name/value pairs, detailed below, or one of the following explicit assignments: spmplot(Y,group);
spmplot(Y,group,plo);
spmplot(Y,group,plo,dispopt);
where group, plo and dispopt have the meaning described in the pairs/values section.
If varargin{1} (that is second input element) is a n-elements vector, then it is interpreted as a grouping variable vector 'group'. In this case, it can only be followed by 'plo' and 'dispopt'. Otherwise, the program expects a sequence of name/value pairs.
If first input Y is a structure (generally created by function FSMeda), then this structure must have the following fields: Required fields in input structure Y.
| Value | Description |
|---|---|
Y |
a data matrix of size n-by-v. If the input structure Y contains just the data matrix, a standard static scatter plot matrix will be created. On the other hand, if Y also contains information on statistics monitored along a search, then the scatter plots will be linked with other (forward) plots with interaction possibilities, enabled via brushing and datatooltip. More precisely, with option databrush it is possible to create an automatic interaction with the other plots, while with option datatooltip it is possible to retrieve information about a particular unit once selected with the mouse). Optional fields in input structure Y. |
MAL |
matrix containing the Mahalanobis distances monitored in each step of the forward search. Every row is associated with a unit (this is a necessary field if the user wants to brush the scatter plot matrix). |
Un |
matrix containing the order of entry of each unit (necessary if datatooltip is true or databrush is not empty). |
label |
cell of length n containing the labels of the units. This optional argument is used in conjuction with options databrush and datatooltip. When datatooltip=1, if this field is not present labels row1, ..., rown will be automatically created and included in the pop up datatooltip window else the labels contained in Y.label will be used. When databrush is a cell and it is called together with option 'labeladd' '1', the trajectories in the malfwdplot will be labelled with the labels contained in Y.label. |
Data Types: single|double
Name-Value Pair Arguments
Specify optional comma-separated pairs of Name,Value arguments.
Name is the argument name and Value
is the corresponding value. Name must appear
inside single quotes (' ').
You can specify several name and value pair arguments in any order as
Name1,Value1,...,NameN,ValueN.
'colorBackground',true
, 'dispopt','box'
, 'group',group
, 'nameY',["aa" "bb" "cc"]
, 'nameYrot',30
, 'nameYlength',3
, 'order','FPC'
, 'overlay',1
, 'plo',true
, 'selunit','3'
, 'tag','myspm'
, 'typespm','lower'
, 'undock', [1 1; 1 3; 3 4]
, 'datatooltip',''
, 'databrush',1
, 'subsize',10:100
, 'selstep',100
colorBackground
—background color of each scatter which depends on the
value of the correlation coefficient.boolean.
If colorBackground is false (default), no color is applied, else a background color which depends on the value of the correlation coefficient is used.
Example: 'colorBackground',true
Data Types: logical
dispopt
—what to put on the diagonal.character.
String which controls how to fill the diagonals in a plot of Y vs Y (main diagonal of the scatter plot matrix). Set dispopt to 'hist' (default) to plot histograms, or 'box' to plot boxplots.
REMARK 1: the style which is used for univariate boxplots is 'traditional' if the number of groups is <=5, else it is 'compact'.
Example: 'dispopt','box'
Data Types: char
group
—grouping variable.vector with n elements.
group is a grouping variable defined as a categorical variable, numeric, or array of strings, or string matrix, and it must have the same number of rows of Y. This grouping variable that determines the marker and color assigned to each point.
Remark: if 'group' is used to distinguish a set of outliers from a set of good units, the id number for the outliers should be the larger (see optional field 'labeladd' of option 'plo' for details).
Example: 'group',group
Data Types: char
nameY
—Vector of length v containing the names of the variables.string array of cell array of characters.
Note that this optional argument is ignored if Y is a table. In this last case the variableNames are used.
Example: 'nameY',["aa" "bb" "cc"]
Data Types: cell array of characters or string array
nameYrot
—Angle (in degrees) for the labels of the variables.the default is 90, which means no rotation with respect to the MATLAB default.
The setting nameYrot = 0 produces a rotation perpendicular to the axes, which is often used in R graphics. Intermediate values are of course possible.
Example: 'nameYrot',30
Data Types: numeric scalar.
nameYlength
—scalar indicating the maximum length of the labels
of the variables.it is used to shorten the labels when they are too long.
The default value is 0, which indicates that the labels length should not be changed.
Example: 'nameYlength',3
Data Types: numeric scalar.
order
—Order of the variables in the scatterplot matrix.character | string.
Order in which the variables are shown inside spmplot. Variable reordering can be used to improve the visualization by placing similar variables next to each other.
Possible values are: "alphabet"=alphabetical order is used;
"original"= the order in the input argument Y is preserved (this is the default option);
"AOE" = the angular order of the first two eigenvectors is used.
More precisely, the order of the variables is calculated from the order of the angles, $a_i$:
\[ a_i= tan^{-1}(e_{i2}/e_{i1}) \qquad \mbox{if} \qquad e_{i1}>0; \] \[ a_i= tan^{-1}(e_{i2}/e_{i1})+\pi \qquad \mbox{if} \qquad e_{i1} \leq 0. \]where $e_1=(e_{11}, \ldots, e_{p1})'$ and $e_2=(e_{12}, \ldots, e_{p2})'$ are the first two eigenvectors associated with the first two largest eigenvalues of the correlation matrix. Additional information can be found in Friendly (2002).
"FPC" = for the first principal component order.
Example: 'order','FPC'
Data Types: char or string
overlay
—Superimposition on the panels out of the main diagonal of
the scatter matrix.scalar, char | structure.
It specifies what to add in the background for the panels specified in undock (default is for all oh them).
The default value is overlay='', i.e. nothing is changed. If overlay=1 the the filled contours are added to each panel, considering all groups, as default. If overlay is a structure it may contain the following fields:
| Value | Description |
|---|---|
type |
Type of plot to add in the background or to superimpose. String. It can be: 'contourf', 'contour', 'ellipse' or 'boxplotb', specifying respectively to add filled contour (default when overlay=1), contour, ellipses or a bivariate boxplot (see function boxplotb.m). |
include |
Boolean vector specifying which groups to include in the type of plot specified in overlay.type, the default value is a vector of ones (i.e. all groups). |
cmap |
The colormap for the type 'contourf' and 'contour' is grey as default. In these case, this field may specify the colors used for the color map. It is a three-column matrix of values in the range [0,1] where each row is an RGB triplet that defines one color. Check the colormap function for additional informations. |
conflev |
When the type specified is 'ellipse', the size of the ellipses is chi2inv(0.95,2) as default. In this case, this field may specify a different confidence level used and it is a value between 0 and 1. |
Example: 'overlay',1
Data Types: single | double
plo
—names, labels, colors, marker type.empty value, scalar | structure.
This options controls the names which are displayed in the margins of the scatter-plot matrix and the labels of the legend.
If plo is the empty vector [], then nameY and labeladd are both set to the empty string '' (default), and no label and no name is added to the plot.
If plo = 1 the names Y1,..., Yv or the variable names of the table are added to the margins of the the scatter plot matrix else nothing is added.
If plo is a structure, it is possible to control not only the names but also, point labels, colors, symbols. More precisely structure pl may contain the following fields:
| Value | Description |
|---|---|
labeladd |
if it is '1', the elements belonging to the max(group) in the spm are labeled with their unit row index or their row name. The row name is taken from plo.label or if plo.label is empty from The default value is labeladd = '', i.e. no label is added. |
nameY |
cell array of strings containing the labels of the variables. As default value, the labels which are added are Y1, ..., Yv. |
nameYrot |
Angle (in degrees) for the labels of the variables. The default is 90, which means no rotation with respect to the MATLAB default. The setting nameYrot = 0 produces a rotation perpendicular to the axes, which is often used in R graphics. Intermediate values are of course possible. |
nameYlength |
scalar indicating the maximum length of the labels of the variables. It is used to shorten the labels when they are too long. The default value is 0, which indicates that the labels length should not be changed. |
clr |
a string of color specifications. By default, the colors are 'brkmgcy'. |
sym |
a string or a cell of marker specifications. For example, if sym = 'o+x', the first group will be plotted with a circle, the second with a plus, and the third with a 'x'. This is obtained with the assignment plo.sym = 'o+x' or equivalently with plo.sym = {'o' '+' 'x'}. By default the sequence of marker types is: '+';'o';'*';'x';'s';'d';'^';'v';'>';'<';'p';'h';'.' plo.siz: scalar, a marker size to use for all plots. By default the marker size depends on the number of plots and the size of the figure window. Default is siz = '' (empty value). plo.doleg: a string to control whether legends are created or not. Set doleg to 'on' (default) or 'off'. plo.label : cell of length n containing the labels of the units. If this field is empty the sequence 1:n will be used to label the units. plo.TickLabels : character array containing the formatSpec applied to the TickLabels of the axes. By default the user can leave this field unspecified, and in this case the formatSpec is determined automatically. Otherwise the user should specify a formatSpec following the standard syntax of sprintf and fprintf, or can set TickLabels to '' or [] to remove the TickLabels from the axes. |
Example: 'plo',true
Data Types: Empty value, scalar or structure.
selunit
—unit labelling in the spmplot and in the malfwdplot.cell array of strings | string | numeric vector for labelling units.
When input argument Y is a structure the threshold is associated with the trajectories of the Mahalanobis distances monitored along the search.
If it is a cell array of strings, only the units that in at least one step of the search had a Mahalanobis distance greater than selunit{1} or smaller than selline{2} will have a textbox in the scatter plot matrix and in the associated malfwdplot after brushing.
If it is a string it specifies the threshold above which labels have to be put. For example selunit='2.6' means that the text labels in the scatter plot matrix (and in the malfwdplot after brushing) are added only for the units which have in at least one step of the search a value of the Mahalanobis distance greater than 2.6.
If it is a numeric vector it contains the list of the units for which it is necessary to put the text labels in each panel of the spmplot and in the associated malfwdplot (if input option databrush is not empty). For example if selunit is [20 34], the labels associated to rows 20 and 34 are added to each scatter plot. The labels which are used are taken from Y.label if Y is a structure or from plo.label if plo.label is not empty and Y is a 2D array, else the numbers 1:n are used.
The default value of selunit is string '2.5' if input argument Y is a structure else it is an empty value if input argument Y is a matrix.
Example: 'selunit','3'
Data Types: numeric or character
tag
—plot tag.string.
string which identifies the handle of the plot which is about to be created. The default is to use tag 'pl_spm'. Notice that if the program finds a plot which has a tag equal to the one specified by the user, then the output of the new plot overwrites the existing one in the same window else a new window is created.
Example: 'tag','myspm'
Data Types: char
typespm
—type of scatter plot matrix.character/string | struct.
If typespm is 'full' (default) panels above and below the main diagonal are shown with scatter. If typespm is 'lower' scatter plots are shown just below the main diagonal. The upper part of the scatter plot matrix contains the values of the correlation coefficients. If optional input argument group is present, the correlation coefficient is also computed for each group and shown in each scatter. The size of the label of the correlation coefficient reflects his magnitude (in absolute value). It typespm is 'upper' the upper part of the scatter plot matrix contains the scatters and the lower part just the values of the correlations.
If typespm is a struct it is possible to control how the lower (upper) part of the scatter plot matrix is shown.
More precisely, typespm.lower controls how the part below the diagonal is shown and typespm.upper controls the upper part. Possible entries of typespm.lower or typespm.upper are "scatter", "circle", "square" "number" (number without color), "cnumber" (colored number) and "none".
For example if
| Value | Description |
|---|---|
lower |
"number" and |
upper |
"circle", the correlations in the part below the diagonal are shown with numbers and the upper part with circles. Similarly, if typespm.lower="none" and |
scatter |
"none" just the upper part of the scatter plot matrix is shown. The fields of the struct can either be characters or strings. typespm='lower' is equivalent to typespm=struct; typespm.lower='scatter', typespm.upper='number'; |
Example: 'typespm','lower'
Data Types: scalar char or string or alternatively a struct
undock
—Panel to undock and visualize separately.matrix | logical matrix.
If undock='' (default), no panel is extracted. If undock is a r-by-2 matrix, it specifies the r coordinates of the scatter plot matrix to undock and visualize separately in a bivariate plot (i.e. for panels out of the main diagonal plots) or in an univariate plot (i.e. the ones on the main diagonal). If undock is a v-by-v logical matrix, where v are the number of columns in Y, the trues of undock are undocked and visualized separately.
REMARK - When used, undock automatically deletes the plots obtained by spmplots. If it is desired to keep some of them, the respective 'Tag' associated has to be changed (e.g.
selecting the figure and then: set(gcf,'Tag','newTag');).
Example: 'undock', [1 1; 1 3; 3 4]
Data Types: single | double | logical
datatooltip
—interactive clicking.empty value (default) | structure.
If datatooltip is not empty the user can use the mouse in order to have information about the unit selected, the step in which the unit enters the search and the associated label. If datatooltip is a structure, it may contain the following fields:
| Value | Description |
|---|---|
DisplayStyle |
Determines how the data cursor displays. |
SnapToDataVertex |
Specifies whether the data cursor snaps to the nearest data value or is located at the actual pointer position. The default options of the structure are DisplayStyle='Window' and SnapToDataVertex='on'. |
Example: 'datatooltip',''
Data Types: empty value, scalar or struct
databrush
—interactive mouse brushing.empty value (default), scalar | cell.
DATABRUSH IS AN EMPTY VALUE.
If databrush is an empty value (default), no brushing is done.
The activation of this option (databrush is a scalar or a cell) enables the user to select a set of observations in the current plot and to see them highlighted in the malfwdplot, i.e. the plot of the trajectories of all observations, grouped according to the selection(s) done by brushing. If the malfwdplot does not exist it is automatically created.
In addition, brushed units can be highlighted in the other following plots (only if they are already open): - minimum Mahalanobis distance plot;
Remark. the window style of the other figures is set equal to that which contains the spmplot. In other words, if the scatterplot matrix plot is docked all the other figures will be docked too.
DATABRUSH IS A SCALAR.
If databrush is a scalar the default selection tool is a rectangular brush and it is possible to brush only once (that is persist='').
DATABRUSH IS A CELL.
If databrush is a cell, it is possible to use all optional arguments of function selectdataFS and the following optional argument: - persist = Persistent brushing.
Persist is an empty value or a scalar containing the strings 'on' or 'off'.
The default value of persist is '', that is brushing is allowed only once.
If persist is 'on' or 'off' brushing can be done as many time as the user requires.
If persist='on' then the unit(s) currently brushed are added to those previously brushed. It is possible, every time a new brushing is done, to use a different color for the brushed units.
If persist='off' every time a new brush is performed units previously brushed are removed.
- labeladd= point labelling. If this option is '1', we label the units of the last selected group with the unit row index in input Y if Y is a matrix or with the labels contained in Y.label if input Y is a struct.
The default value is labeladd='', i.e. no label is added in the malfwdplot.
Remark: The options which follow (subsize, selstep and selunit) work in connection with previous option databrush and produce their effect on the monitoring MD plot (malfwdplot). Note that the options which follow can only be used if the first argument of spmplot is a structure containing information about the fwd search (i.e. the fields MAL, Un and eventually label).
Example: 'databrush',1
Data Types: single | double | struct
subsize
—x axis control in malfwdplot.vector.
numeric vector containing the subset size with length equal to the number of columns of matrix Y.MAL.
If it is not specified it will be set equal to size(Y.MAL,1)-size(Y.MAL,2)+1:size(Y.MAL,1)
Example: 'subsize',10:100
Data Types: single | double
selstep
—position of text labels of brushed units in malfwdplot.vector.
Numeric vector which specifies for which steps of the forward search text labels are added in the monitoring MD plot after a brushing action in the spmplot. The default is to write the labels at the initial and final step. The default is selstep=[m0 n] where m0 and n are respectively the first and final step of the search.
Example: 'selstep',100
Data Types: single | double
Output Arguments
BigAx —handle to big (invisible) axes framing the subaxes.
Scalar
See gplotmatrix for further details.
More About
Additional Details
spmplot has the same output of gplotmatrix in the statistics toolbox: [H,AX,BigAx] = spmplot(...) returns an array of handles H to the plotted points; a matrix AX of handles to the individual subaxes; and a handle BIGAX to big (invisible) axes framing the subaxes. The third dimension of H corresponds to groups in G. AX contains one extra row of handles to invisible axes in which the histograms are plotted. BigAx is left as the CurrentAxes so that a subsequent TITLE, XLABEL, or YLABEL will be centered with respect to the matrix of axes.
References
Friendly M. (2002), Corrgrams: Exploratory Displays for Correlation Matrices. The American Statistician, v. 56, pp. 316–324, https://doi.org/10.1198/000313002533