mdpattern
mdpattern finds and plots missing data patterns
Syntax
Description
Example of the use of options dispresults and plots.Mispat
=mdpattern(Y,
Name, Value)
[
Example of mdpattern with timetable input.Mispat,
tMisAndOut]
=mdpattern(___)
Examples
 mdpattern with table input.
mdpattern with table input.
 mdpattern with table input.
mdpattern with table input.Load the nhanes data The nhanes data is a dataset with 25 observations on the following 4 variables.
% age, Age group (1=20-39, 2=40-59, 3=60+).
% bmi, Body mass index (kg/m**2).
% hyp, Hypertensive (1=no,2=yes).
% chl, Total serum cholesterol (mg/dL).
% namvar array of strings containing the names of the columns of X.
namvar=["age" "bmi" "hyp" "chl"];
X=[1 NaN NaN NaN
2 22.7 1 187
1 NaN 1 187
3 NaN NaN NaN
1 20.4 1 113
3 NaN NaN 184
1 22.5 1 118
1 30.1 1 187
2 22.0 1 238
2 NaN NaN NaN
1 NaN NaN NaN
2 NaN NaN NaN
3 21.7 1 206
2 28.7 2 204
1 29.6 1 NaN
1 NaN NaN NaN
3 27.2 2 284
2 26.3 2 199
1 35.3 1 218
3 25.5 2 NaN
1 NaN NaN NaN
1 33.2 1 229
1 27.5 1 131
3 24.9 1 NaN
2 27.4 1 186];
Xtable=array2table(X,VariableNames=namvar);
[Mispat,tMisAndOut]=mdpattern(Xtable);
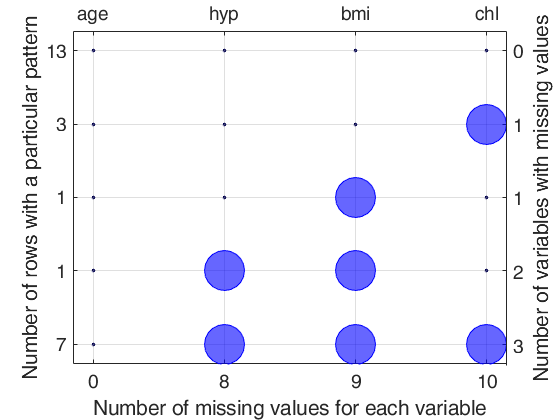
Detailed explanation of the "Missing data pattern figure" Top axis contains the names of the variables. Big circle means missing value; smaller filled dot represents non missing value. Left axis shows the number of observations for each pattern For example number 13 shows that the associated pattern is repeated 13 times. The sum of the numbers on the left axis is n, the total number of rows. Right axis counts the variables with missing values and it is equal to the number of big circles in the corresponding row. The number of missing values for each variable is shown on the bottom axis.
 Example of the use of options dispresults and plots.
Example of the use of options dispresults and plots.
 Example of the use of options dispresults and plots.
Example of the use of options dispresults and plots.Load the nhanes data The nhanes data is a dataset with 25 observations on the following 4 variables.
% age, Age group (1=20-39, 2=40-59, 3=60+).
% bmi, Body mass index (kg/m**2).
% hyp, Hypertensive (1=no,2=yes).
% chl, Total serum cholesterol (mg/dL).
% namvar array of strings containing the names of the columns of X.
namvar=["age" "bmi" "hyp" "chl"];
X=[1 NaN NaN NaN
2 22.7 1 187
1 NaN 1 187
3 NaN NaN NaN
1 20.4 1 113
3 NaN NaN 184
1 22.5 1 118
1 30.1 1 187
2 22.0 1 238
2 NaN NaN NaN
1 NaN NaN NaN
2 NaN NaN NaN
3 21.7 1 206
2 28.7 2 204
1 29.6 1 NaN
1 NaN NaN NaN
3 27.2 2 284
2 26.3 2 199
1 35.3 1 218
3 25.5 2 NaN
1 NaN NaN NaN
1 33.2 1 229
1 27.5 1 131
3 24.9 1 NaN
2 27.4 1 186];
Xtable=array2table(X,VariableNames=namvar);
% Plot is not shown.
plots=false;
% option dispresults shows a detailed explanation of
% the content of two output matrices in the command window.
dispresults=true;
[Mispat,tMisAndOut]=mdpattern(Xtable,'plots',false,'dispresults',dispresults);
Table which shows missing values patterns
NrowsWithPattern age hyp bmi chl NvarWithMis
________________ ____ ____ ____ _____ ___________
Pattern1 13.00 1.00 1.00 1.00 1.00 0.00
Pattern2 3.00 1.00 1.00 1.00 0.00 1.00
Pattern3 1.00 1.00 1.00 0.00 1.00 1.00
Pattern4 1.00 1.00 0.00 0.00 1.00 2.00
Pattern5 7.00 1.00 0.00 0.00 0.00 3.00
totPatOrMis 25.00 0.00 8.00 9.00 10.00 27.00
0 means missing value and 1 represents non missing value
First column contains the number of observations for each pattern
For example number 13 shows that the associated pattern is repeated 13 times
The sum of the numbers in the first column is n, that is the total number of rows
The last column shows the number of variables with missing values for that particular pattern
------------------------
Missing value and outlier report
Mean Median Stdev MAD Count_miss Perc_miss outInf outSup
______ ______ _____ _____ __________ _________ ______ ______
age 1.76 2.00 0.83 1.48 0.00 0.00 0.00 0.00
bmi 26.56 26.75 4.22 4.60 9.00 36.00 0.00 0.00
hyp 1.24 1.00 0.44 0.00 8.00 32.00 0.00 4.00
chl 191.40 187.00 45.22 28.17 10.00 40.00 3.00 1.00
Columns outInf and outSup contain the number of units which are
above x0.75+1.5*IQR or below x0.25-1.5*IQR, where IQR is the interquartile range
Example of mdpattern with timetable input.
TT = readtimetable('outages.csv');
[A,B]=mdpattern(TT(:,["Loss" "Customers" ]))
Related Examples
 An example with 2 simulated patterns of missing values.
An example with 2 simulated patterns of missing values.
 An example with 2 simulated patterns of missing values.
An example with 2 simulated patterns of missing values.
close all
n=10000;
p=10;
X=randn(n,p);
% Create first missing data pattern
n1=300; n2=3;
rowsWithMis=randsample(n,n1);
colsWithMis=randsample(p,n2);
X(rowsWithMis,colsWithMis)=NaN;
% Create second missing data pattern
n1=120; n2=5;
rowsWithMis=randsample(n,n1);
colsWithMis=randsample(p,n2);
X(rowsWithMis,colsWithMis)=NaN;
mdpattern(X);
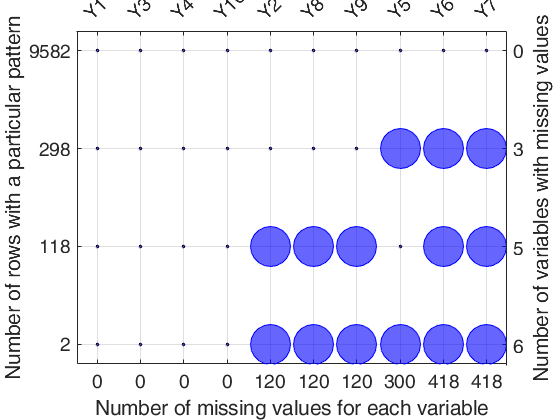
Detailed explanation of the "Missing data pattern figure" Top axis contains the names of the variables. Big circle means missing value; smaller filled dot represents non missing value. Left axis shows the number of observations for each pattern For example number 9582 shows that the associated pattern is repeated 9582 times. The sum of the numbers on the left axis is n, the total number of rows. Right axis counts the variables with missing values and it is equal to the number of big circles in the corresponding row. The number of missing values for each variable is shown on the bottom axis.
Input Arguments
Y — data matrix (2D array) table or timetable containing
observations on v quantitative variables.
Data Types - matrix, table or timetable.
Data Types: single| double
Name-Value Pair Arguments
Specify optional comma-separated pairs of Name,Value arguments.
Name is the argument name and Value
is the corresponding value. Name must appear
inside single quotes (' ').
You can specify several name and value pair arguments in any order as
Name1,Value1,...,NameN,ValueN.
'Lc',{'Y1' Y2' 'Y3' 'Y4'}
, 'plots',false
, 'dispresults',true
Lc
—Vector of column labels.cell array of charaters | String array.
Lc contains the labels of the columns of the input array Y. This option is unnecessary if Y is a table, because in this case Lc=X.Properties.VariableNames;
Example: 'Lc',{'Y1' Y2' 'Y3' 'Y4'}
Data Types: cell array of characters or String.
plots
—Plot on the screen.boolean If plots = true (default), a plot which displays missing data patterns is displayed on the screen.
Top axis contains the name of the variables Big circle means missing value; smaller filled dot represents non missing value Left axis shows the number of observations for each pattern. For example number 40 shows that the associated patterns is repeated 40 times.
The sum of the numbers on the left axis is n the total number of rows Right axis counts the variables with missing values and it is equal to the number of big circles in the corresponding row.
Example: 'plots',false
Data Types: Boolean
dispresults
—Display results on the screen.boolean.
If dispresults is true it is possible to see on the screen the two output tables Xpat,tMisAndOut.
The default value of dispresults is false.
Example: 'dispresults',true
Data Types: Boolean
Output Arguments
Mispat —missing values pattern.
table
table with size (k+1)x(v+2), where k is the total number of missing values patterns which are present in the data matrix. The first k rows contain the patterns.
The last row contains n and then the total number of missing values in each column. The first column contains information about the number of observations for each pattern.
The columns of Mispat are sorted in non decreasing number of outliers. The last column contains the number of variables with missing values for each pattern.
tMisAndOut —missing values and univariate outliers for each variable.
The rows of this table are associated with the variables.
The columns are referred to a series of statistics.
More precisely: Columns 1:4 contain mean and median, std deviation and rescaled MAD (median absolute deviation).
Fifth column (Count_miss) contains the number of missing values for each variable.
Sixth column (Percmiss) conatins the percentage of missing data for each variable.
Seventh and eight column contain the number of outliers respectively in the left and right tail of the distribution. The criterion to decide whether a unit is outlier is based on the boxplot concept, that is the outliers are the units which are above x0.75+1.5*IQR or below x0.25-1.5*IQR, where IQR is the interquartile range.
References
Schafer, J.L. (1997). "Analysis of Incomplete Multivariate Data". London: Chapman & Hall.