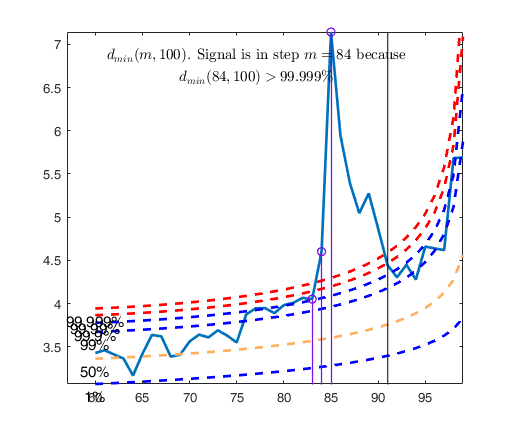
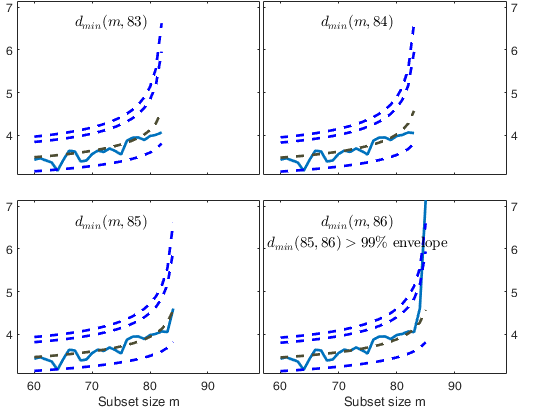
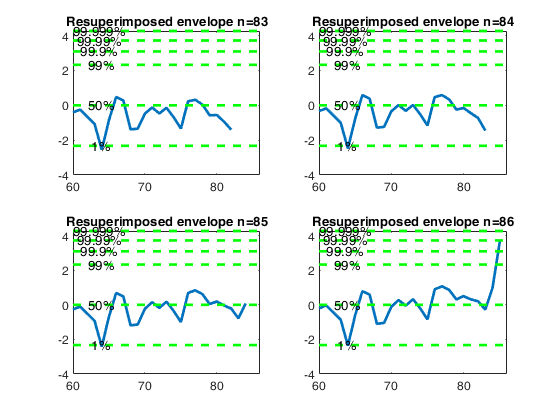
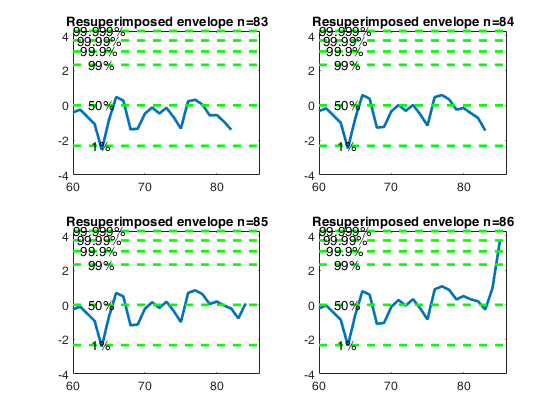
Let and d_{\mbox{min}}(m) be respectively the deletion
distance for unit i based
on a subset of size m and d_{\mbox{min}}(m) the min. Mahalanobis
distance in the forward search at step m. Testing for outliers requires a
reference distribution for d^2_i(m ) in and hence for
d_{\mbox{min}}(m) in (\ref{min}). When \Sigma is estimated from all
n observations, the squared statistics have an F distribution.
However, the estimate \hat{\Sigma}(m) in the search uses the central
m out of n observations, so that the variability is underestimated.
The consistency factor c(m,n) given below
c(m,n)=\frac{n}{m} C_{v+2} \{C_{v}^{-1} (m/n) \}
where C_r is the c.d.f. of the \chi^2 distribution on r degrees of
freedom, allows for estimation from this truncated distribution, providing an
approximately unbiased estimate of \Sigma.
We can treat the distribution of the rescaled deletion Mahalanobis distance
c(m,n)d_{\mbox{min}}^2(m) as a squared deletion distance on m-1
degrees of freedom, whose distribution is (Atkinson Riani and Cerioli,
2004; pp. 43-44)
\begin{equation}\label{F}
\frac{m^2-1}{m(m-v)} F_{v,m-v},
\end{equation}
The distribution of the rescaled min Mahalanobis distance
c(m,n) d_{\mbox{min}}^2(m)
of a subset of size m constructed in such a way that
the centroid and covariance matrix of the subset are taken using the
units having the m smallest Mahalanobis distances can be treated as
the distribution of the (m+1)th order statistic from (F_{v,m-v}).
The results of order statistics Y_{(1)}, Y_{(2)}, \cdots, Y_{(n)} from a sample of size n from a distribution with CDF G(y), state that
\begin{equation}
\label{orderstat}
P\{Y_{(m+1)} \le y \} = P \left\{ F_{2(n-m),2(m+1)} > \frac{1-G(y)}{G(y)} \times
\frac{m+1}{n-m} \right\}
\end{equation}
Given that in our case G(y) is the CDF of the F_{v,m-v} we can rewrite this equation as
\begin{eqnarray*}
&& P\{d_{\mbox{ min}}^2(m) \leq \widehat{ d_{\mbox{min}}^2(m)} \} = \\
&& 1- F_{2(n-m),2(m+1)} \left( \left( \frac{1}{ F_{v,m-v} \left( \frac{m(m-v)}{m^2-1 } c(m,n) d_{\mbox{min}}^2(m) \right) }-1 \right) \frac{m+1}{n-m} \right)
\end{eqnarray*}
where F_{a,b}(y) is the CDF of the F distribution with a and b degrees of freedom evaluated in y.
The value of the min. Mahalanobis distance transformed in normal coordinates computed by this routine is
nothing but
\Phi^{-1} \left( P\left\{ d_{\mbox{min}}^2(m) \leq \widehat{ d_{\mbox{min}}^2(m)} \right\} \right)
where \Phi^{-1} is the inverse of the CDF of the standard normal distribution.


 Resuperimposing envelopes and normal coordinates.
Resuperimposing envelopes and normal coordinates.