fanplotFS
fanplotFS plots the fan plot for transformation in linear regression or deletion t stat
Description
fanplotFS with all default options.brushedUnits
=fanplotFS(out)
fanplotFS with optional arguments.brushedUnits
=fanplotFS(out,
Name, Value)
Examples
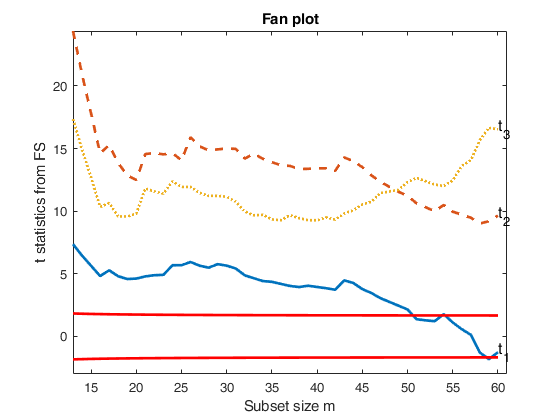
 fanplotFS with all default options.
fanplotFS with all default options.
 fanplotFS with all default options.
fanplotFS with all default options.load the wool data
XX=load('wool.txt');
y=XX(:,end);
X=XX(:,1:end-1);
% FSRfan and fanplotFS with all default options
[out]=FSRfan(y,X);
fanplotFS(out);Total estimated time to complete LMS: 0.05 seconds ------------------------------ Warning: Number of subsets without full rank equal to 15.3% Total estimated time to complete LMS: 0.09 seconds ------------------------------ Warning: Number of subsets without full rank equal to 15.3% Total estimated time to complete LMS: 0.01 seconds ------------------------------ Warning: Number of subsets without full rank equal to 15.3% Total estimated time to complete LMS: 0.01 seconds ------------------------------ Warning: Number of subsets without full rank equal to 15.3% Total estimated time to complete LMS: 0.01 seconds ------------------------------ Warning: Number of subsets without full rank equal to 15.3%
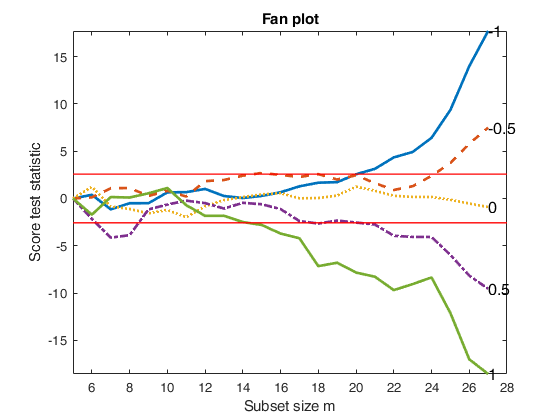
 fanplotFS with optional arguments.
fanplotFS with optional arguments.
 fanplotFS with optional arguments.
fanplotFS with optional arguments.FSRfan and fanplotFS with specified lambda
load('loyalty.txt');
y=loyalty(:,4);
X=loyalty(:,1:3);
% la = vector contanining the most common values of the transformation
% parameter
la=[-1 -0.5 0 0.5 1];
[out]=FSRfan(y,X,'la',la);
fanplotFS(out);Total estimated time to complete LMS: 0.02 seconds Total estimated time to complete LMS: 0.01 seconds Total estimated time to complete LMS: 0.02 seconds Total estimated time to complete LMS: 0.02 seconds Total estimated time to complete LMS: 0.02 seconds
Related Examples
Interactive example 1.
FSRfan and fanplotFS with databrush option.
load('loyalty.txt');
y=loyalty(:,4);
X=loyalty(:,1:3);
la=[-1 -0.5 0 0.5 1];
[out]=FSRfan(y,X,'la',la);
fanplotFS(out,'databrush','1');
Interactive example 2.
FSRfan and fanplotFS with databrush, persist, label and RemoveLabels options.
%Removelabels is a parameter of SelectdataFS function
load('loyalty.txt');
y=loyalty(:,4);
X=loyalty(:,1:3);
la=[-1 -0.5 0 0.5 1];
[out]=FSRfan(y,X,'la',la);
fanplotFS(out,'databrush',{ 'persist' 'on' 'Label' 'on' 'RemoveLabels' 'off'});
Interactive example 3.
FSRfan and fanplotFS with databrush, bivarfit, label and RemoveLabels options.
load('loyalty.txt');
y=loyalty(:,4);
X=loyalty(:,1:3);
la=[-1 -0.5 0 0.5 1];
[out]=FSRfan(y,X,'la',la);
fanplotFS(out,'databrush',{ 'bivarfit' '2' 'Label' 'on' 'RemoveLabels' 'off'});
Interactive example 4.
FSRfan and fanplotFS with databrush and selectionmode options.
%Example of the use of persistent cumulative brush.
%Every time a brushing action is performed
%current highlightments are added to previous highlightments
load('loyalty.txt');
y=loyalty(:,4);
X=loyalty(:,1:3);
la=[-1 -0.5 0 0.5 1];
[out]=FSRfan(y,X,'la',la);
fanplotFS(out,'databrush',{'selectionmode','Brush'});
fanplotFS(out,'databrush',{'selectionmode' 'Lasso' 'persist' 'off'})
fanplotFS(out,'databrush',{'selectionmode' 'Rect' 'persist' 'on'})
fanplotFS with datatooltip passed as scalar.
That is using default options for datacursor (i.e. DisplayStyle=window).
load('loyalty.txt');
y=loyalty(:,4);
X=loyalty(:,1:3);
la=[-1 -0.5 0 0.5 1];
[out]=FSRfan(y,X,'la',la);
fanplotFS(out,'datatooltip',1);
Construct fan plot specifying the confidence level and the xlimits.
load('loyalty.txt');
y=loyalty(:,4);
X=loyalty(:,1:3);
la=[0 1/3 0.4 0.5];
[out]=FSRfan(y,X,'la',la,'init',size(X,2)+2,'nsamp',20000);
fanplotFS(out,'xlimx',[100 300],'conflev',0.95);
Interactive example 5.
Example of the use of multivarfit and xlimx.
load('loyalty.txt');
y=loyalty(:,4);
X=loyalty(:,1:3);
la=[0 1/3 0.4 0.5];
[outs]=FSRfan(y,X,'la',la,'init',size(X,2)+2,'nsamp',20000);
fanplotFS(outs,'xlimx',[10 520],'databrush',{'selectionmode' 'Brush' 'multivarfit' '2'})
Interactive example 6.
Example of the use of FlagSize, namey, namex, lwd,FontSize, SizeAxesNum.
load('loyalty.txt');
y=loyalty(:,4);
X=loyalty(:,1:3);
la=[-1 -0.5 0 0.5 1];
[out]=FSRfan(y,X,'la',la);
namey='Sales';
nameX={'Number of visits', 'Age', 'Number of persons in the family'};
%FlagSize controls how large must be the highlighted points. It is a
%parametr of selectdataFS.
fanplotFS(out,'xlimx',[10 520],'lwd',1.5,'FontSize',11,'SizeAxesNum',11,'nameX',nameX,'namey',namey,'databrush',{'selectionmode' 'Brush'...
'multivarfit' '2' 'FlagSize' '5'})
Interactive example 7.
Only one brush specifying labels for y and X.
load loyalty.mat
y=loyalty{:,4};
X=loyalty{:,1:3};
la=[-1 -0.5 0 0.5 1];
[out]=FSRfan(y,X,'la',la);
namey=loyalty.Properties.VariableNames{4};
nameX=loyalty.Properties.VariableNames(1:3);
fanplotFS(out,'databrush',{'selectionmode' 'Brush' 'FlagSize' '5'},'nameX',nameX,'namey',namey)
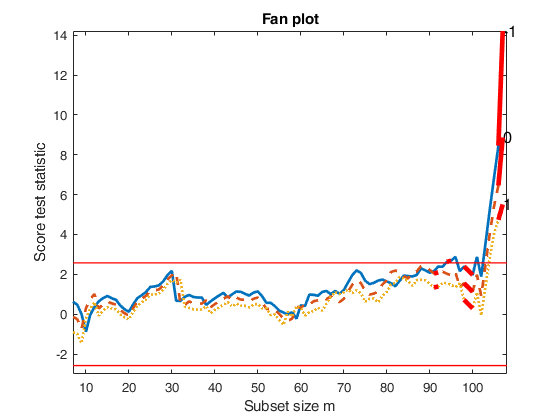
fanplotFS with option highlight (first example).
load the wool data
XX=load('wool.txt');
y=XX(:,end);
X=XX(:,1:end-1);
% FSRfan and fanplotFS with all default options
[out]=FSRfan(y,X);
% Units 24 27 and 23 enter the last three steps in the search with la=0.
% Option highlight enables us to understand when these 3 units join the
% subset for the other values of lambda
fanplotFS(out,'highlight',[24 27 23]);
 fanplotFS with option highlight (second example).
fanplotFS with option highlight (second example).
 fanplotFS with option highlight (second example).
fanplotFS with option highlight (second example).
load gasoline.mat
y=gasoline{:,2};
X=gasoline{:,1};
[out]=FSRfan(y,X,'la', [-1 0 1]);
% In the code below, option highlight enables us to understand when units
% [21 18 33 77] join the subset for the difference values of lambda
fanplotFS(out,'highlight',[ 21 18 33 77]);Total estimated time to complete LMS: 0.01 seconds
Total estimated time to complete LMS: 0.00 seconds
Total estimated time to complete LMS: 0.00 seconds
Steps of entry of selected units when la= -1
95 21
99 18
100 33
107 77
Steps of entry of selected units when la= 0
92 21
99 18
100 33
107 77
Steps of entry of selected units when la= 1
92 21
99 18
100 33
107 77
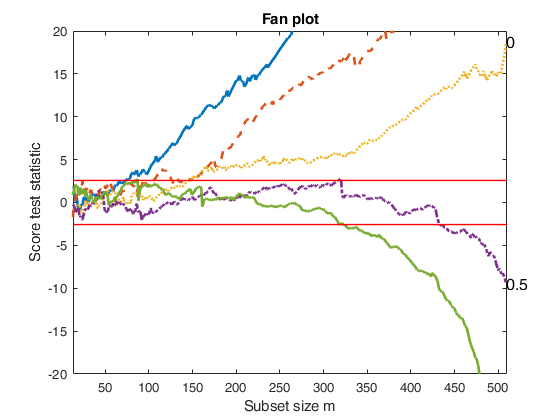
Example of option highlight when highlight is a 2D array.
load('loyalty.txt');
y=loyalty(:,4);
X=loyalty(:,1:3);
% la = vector contanining the most common values of the transformation
% parameter lambda
la=[0 0.5 1];
[outFSRfan]=FSRfan(y,X,'la',la);
% Store the units declares as outliers for the 3 values of lambda
Highl=NaN(250,5);
for j=1:length(la)
if abs(la(j))>1e-06
outlaj=FSR(y.^la(j),X,'msg',0,'plots',0);
else
outlaj=FSR(log(y),X,'msg',0,'plots',0);
end
oultlj=outlaj.outliers;
Highl(1:length(oultlj),j)=oultlj;
end
% Highlight in red inside the fanplot the steps of entry of the units
% declared as outliers for the different trajectories of lambda
fanplotFS(outFSRfan,'highlight',Highl,'ylimy',[-50 20],'xlimx',[30 510]);
 fanplotFS based on the output of FSRaddt.
fanplotFS based on the output of FSRaddt.
 fanplotFS based on the output of FSRaddt.
fanplotFS based on the output of FSRaddt.
n=200;
p=3;
randn('state', 123456);
X=randn(n,p);
% Uncontaminated data
y=randn(n,1);
nameX={'F1','F2','F3'};
[out]=FSRaddt(y,X,'plots',0);
% out.la contains the names of the variables which have to be shown
out.la=nameX;
fanplotFS(out);Total estimated time to complete LMS: 0.01 seconds Total estimated time to complete LMS: 0.01 seconds Total estimated time to complete LMS: 0.01 seconds
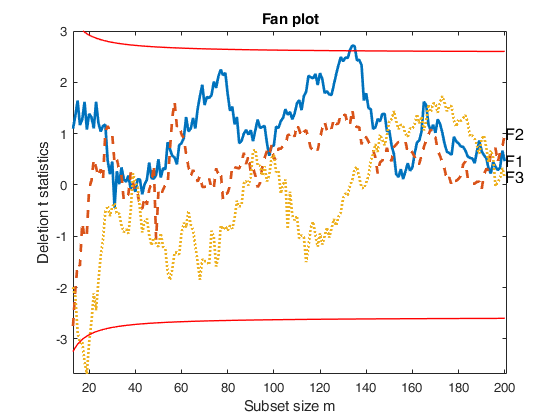
 fanplotFS based on the output of Sregeda.
fanplotFS based on the output of Sregeda.
 fanplotFS based on the output of Sregeda.
fanplotFS based on the output of Sregeda.
load('multiple_regression.txt');
y=multiple_regression(:,4);
X=multiple_regression(:,1:3);
% Specify the type of estimator of t statistic
covrob=2;
% Specify the rho function
rhofunc='optimal';
% Specify line width of the envelopes
lwdenv=2;
[outS]=Sregeda(y,X,'covrob',covrob,'rhofunc',rhofunc);
fanplotFS(outS,'conflev',0.90,'lwdenv',lwdenv);Total estimated time to complete S estimate: 1.05 seconds Total estimated time to complete S estimate: 0.36 seconds Total estimated time to complete S estimate: 0.24 seconds Total estimated time to complete S estimate: 0.24 seconds Total estimated time to complete S estimate: 0.23 seconds Total estimated time to complete S estimate: 0.27 seconds Total estimated time to complete S estimate: 0.20 seconds Total estimated time to complete S estimate: 0.21 seconds Total estimated time to complete S estimate: 0.19 seconds Total estimated time to complete S estimate: 0.20 seconds Total estimated time to complete S estimate: 0.19 seconds Total estimated time to complete S estimate: 0.22 seconds Total estimated time to complete S estimate: 0.23 seconds Total estimated time to complete S estimate: 0.23 seconds Total estimated time to complete S estimate: 0.15 seconds Total estimated time to complete S estimate: 0.15 seconds Total estimated time to complete S estimate: 0.15 seconds Total estimated time to complete S estimate: 0.15 seconds Total estimated time to complete S estimate: 0.14 seconds Total estimated time to complete S estimate: 0.13 seconds Total estimated time to complete S estimate: 0.14 seconds Total estimated time to complete S estimate: 0.15 seconds Total estimated time to complete S estimate: 0.12 seconds Total estimated time to complete S estimate: 0.12 seconds Total estimated time to complete S estimate: 0.12 seconds Total estimated time to complete S estimate: 0.12 seconds Total estimated time to complete S estimate: 0.11 seconds Total estimated time to complete S estimate: 0.11 seconds Total estimated time to complete S estimate: 0.10 seconds Total estimated time to complete S estimate: 0.13 seconds Total estimated time to complete S estimate: 0.13 seconds Total estimated time to complete S estimate: 0.10 seconds Total estimated time to complete S estimate: 0.10 seconds Total estimated time to complete S estimate: 0.11 seconds Total estimated time to complete S estimate: 0.11 seconds Total estimated time to complete S estimate: 0.11 seconds Total estimated time to complete S estimate: 0.09 seconds Total estimated time to complete S estimate: 0.09 seconds Total estimated time to complete S estimate: 0.09 seconds Total estimated time to complete S estimate: 0.09 seconds Total estimated time to complete S estimate: 0.09 seconds Total estimated time to complete S estimate: 0.08 seconds Total estimated time to complete S estimate: 0.07 seconds Total estimated time to complete S estimate: 0.07 seconds Total estimated time to complete S estimate: 0.07 seconds Total estimated time to complete S estimate: 0.07 seconds Total estimated time to complete S estimate: 0.07 seconds Total estimated time to complete S estimate: 0.07 seconds Total estimated time to complete S estimate: 0.07 seconds Total estimated time to complete S estimate: 0.07 seconds
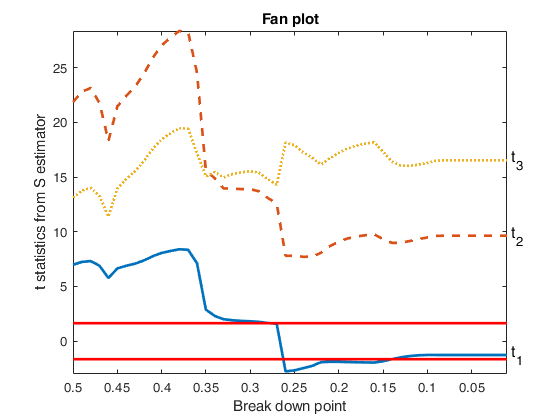
 fanplotFS based on the output of MMregeda.
fanplotFS based on the output of MMregeda.
 fanplotFS based on the output of MMregeda.
fanplotFS based on the output of MMregeda.
load('multiple_regression.txt');
y=multiple_regression(:,4);
X=multiple_regression(:,1:3);
% Specify the type of estimator of t statistic
covrob=5;
% Specify the rho function
rhofunc='optimal';
% Specify line width of the envelopes
lwdenv=2;
[outMM]=MMregeda(y,X,'covrob',covrob,'rhofunc',rhofunc);
fanplotFS(outMM,'conflev',0.90,'lwdenv',lwdenv);Total estimated time to complete S estimate: 0.04 seconds
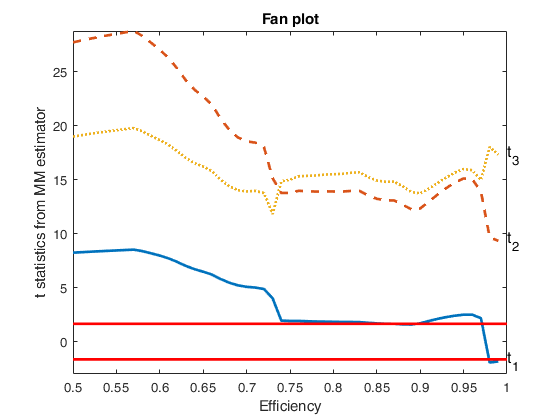
Example of fanplot sent to a subplot (option h).
load('multiple_regression.txt');
y=multiple_regression(:,4);
X=multiple_regression(:,1:3);
% Comparing the monitoring of t tstas using the optimal rho function and
% two different specifications for the covariance matrix of estimates of
% robust regression coefficients
outOPT=Sregeda(y,X,'plots',0,'rhofunc','optimal','covrob',0,'msg',0);
outOPT1=Sregeda(y,X,'plots',0,'rhofunc','optimal','covrob',1,'msg',0);
% Top panel
h=subplot(2,1,1);
fanplotFS(outOPT,'conflev',0.95,'tag','plrobcopv0','h',h);
% Bottom panel
h=subplot(2,1,2);
fanplotFS(outOPT1,'conflev',0.95,'tag','plrobcopv1','h',h);Input Arguments
out — Data to plot.
Structure.
Structure containing the following fields
| Value | Description |
|---|---|
Score |
(n-init) x length(la)+1 matrix: 1st col = fwd search index or bpd or eff 2nd col = value of the statistic in each step of the fwd search or each bdp or each eff, For example if for la(1); ...; last col = value of the score test in each step of the fwd search for la(end). Remark: note that out.Score can be replaced by any other statistic monitored as function of breakdown point, efficiency or subset szie out.Tdel if the input comes from routine FSRaddt. |
la |
vector containing the values of transformation parameter lambda which have been used inside routine FSRfan or the numbers associated to the columns of matrix out.X for which deletion t stats have computed by routine FSRaddt. Alternatively out.la can be a string array or cell arry of characters containing the names of the variables associated with the deletion t stats. |
bs |
matrix of size p x length(la) containing the units forming the initial subset for each value of lambda. out. Un = cell of size length(la). out.Un{i} is a (n-init) x 11 matrix which contains the unit(s) included in the subset at each step of the fwd search (necessary only if option datatooltip or databrush are not empty). |
y |
a vector containing the response (necessary only if option databrush is true). |
X |
a matrix containing the explanatory variables (necessary only if option databrush is not empty). |
Data Types: struct
Name-Value Pair Arguments
Specify optional comma-separated pairs of Name,Value arguments.
Name is the argument name and Value
is the corresponding value. Name must appear
inside single quotes (' ').
You can specify several name and value pair arguments in any order as
Name1,Value1,...,NameN,ValueN.
'addxline', [30 50 200]
, 'conflev',[0.9 0.95 0.99]
, 'flabstep',[20 30 120]
, 'FontSize',12
,'h',h1 where h1=subplot(2,1,1)
, 'highlight',[2 4 20]
, 'corr',1
, 'labx','Subset size m'
, 'laby','Score test statistic'
, 'lwd',2
, 'lwdenv',1
, 'nameX',''
, 'namey',''
, 'multiPanel',true
, 'SizeAxesNum',10
, 'tag','pl_mycov'
, 'titl','Fan plot'
, 'xlimx',[init n]
, 'ylimy',[0 100]
, 'datatooltip',1
, 'databrush',1
addxline
—add to the plot a set of vertical lines.numeric vector.
It is possible to add to the overall plot or to each panel vertical line(s) with constant x-value. The default value is [], that is no vertical line is added.
Example: 'addxline', [30 50 200]
Data Types: double
conflev
—Confidence level.scalar | vector.
Confidence level for the bands (default is 0.99, that is we plot two horizontal lines in correspondence of value -2.58 and 2.58).
Example: 'conflev',[0.9 0.95 0.99]
Data Types: double
flabstep
—numeric vector which specifies the steps of
where to put labels for the trajectories of the
quantities which are monitored.the default is to put the labels in the final steps of the monitoring procedure.
flabstep=[] means no label.
Example: 'flabstep',[20 30 120]
Data Types: double
FontSize
—Font size of the labels.scalar.
Scalar which controls the font size of the labels of the axes and of the labels inside the plot. Default value is 12.
Example: 'FontSize',12
Data Types: double
h
—the axis handle of a figure where to send fanplotFS.axis handle.
This option be used to host the fanplotFS in a subplot of a complex figure formed by different panels
Example: 'h',h1 where h1=subplot(2,1,1)
Data Types: Axes object (supplied as a scalar)
highlight
—units to highlight in the fanplot plot.vector | 2D array | empty (default).
If highlight is a vector it contains numbers associated to the units whose entry step has to be shown in trajectories of the fanplot. More specifically, the steps in which the units inside highlight join the subset is shown in red in the trajectories of the fanplot. The default of highlight is the empty vector []. If highlight is a matrix it contains in column j the units whose entry step has to be shown in the j-th trajectory of the fanplot.
Example: 'highlight',[2 4 20]
Data Types: double
label
—Labels.cell array of strings.
Cell containing the labels of the units (optional argument used when datatooltip=1). If this field is not present labels row1, ..., rown will be automatically created and included in the pop up datatooltip window.
Example: 'corr',1
Data Types: Cell array of strings
labx
—x-axis label.string.
A label for the x-axis (default: 'Subset size m').
Example: 'labx','Subset size m'
Data Types: char or string
laby
—y-axis label.string.
a label for the y-axis (default:'Score test statistic').
Example: 'laby','Score test statistic'
Data Types: char or string
lwd
—Linewidth.scalar.
Scalar which controls linewidth of the curves which contain the score test.
Default line width=2.
Example: 'lwd',2
Data Types: double
lwdenv
—Width of the envelope lines.scalar.
Scalar which controls the width of the lines associated with the envelopes. Default is lwdenv=1.
Example: 'lwdenv',1
Data Types: double
nameX
—Labels of the X variables.cell array of strings.
Cell array of strings of length p containing the labels of the varibles of the regression dataset. If it is empty (default) the sequence X1, ..., Xp will be created automatically.
Example: 'nameX',''
Data Types: Cell array of strings
namey
—Labels of the y variable.string.
String containing the label of the response variable.
Example: 'namey',''
Data Types: char
multiPanel
—plots on a single or multiple panels.boolean.
If multiPanel is true each trajectory appears on a separate subplot. If multiPanel is false (default) all the trajectories appear in single plot. Note that if option multiPanel is supplied option h is ignored.
Example: 'multiPanel',true
Data Types: logical
SizeAxesNum
—Size of the numbers of the axis.scalar.
Scalar which controls the size of the numbers of the axes.
Default value is 10.
Example: 'SizeAxesNum',10
Data Types: double
tag
—Handle of the plot.string.
String which identifies the handle of the plot which is about to be created. The default is to use tag pl_fan. Notice that if the program finds a plot which has a tag equal to the one specified by the user, then the output of the new plot overwrites the existing one in the same window else a new window is created.
Example: 'tag','pl_mycov'
Data Types: char
titl
—Title.string.
A label for the title (default: 'Fan plot')
Example: 'titl','Fan plot'
Data Types: char or string
xlimx
—Min and Max of the x axis.vector.
Vector with two elements controlling minimum and maximum of the x axis.
Default value is [init n].
Example: 'xlimx',[init n]
Data Types: double
ylimy
—Min and Max of the y axis.vector.
Vector with two elements controlling minimum and maximum of the y axis.
Default value for ylimy(1)=max(min(score_test),-20). Default value for ylimy(2)=min(max(score_test),20).
Example: 'ylimy',[0 100]
Data Types: double
datatooltip
—Information about the unit selected.empty value, scalar | structure.
The default is datatooltip=''. If datatooltip is not empty the user can use the mouse in order to have information about the unit selected, the step in which the unit enters the search and the associated label. If datatooltip is a structure, it is possible to control the aspect of the data cursor (see function datacursormode for more details or the examples below). The default options of the structure are DisplayStyle='Window' and SnapToDataVertex='on'.
Example: 'datatooltip',1
Data Types: Empty value, scalar or structure
databrush
—Databrush options.empty value, scalar | cell.
DATABRUSH IS AN EMPTY VALUE: If databrush is an empty value (default), no brushing is done. The activation of this option (databrush is a scalar or a cell) enables the user to select a set of trajectories in the current plot and to see them highlighted in the y|X plot (notice that if the plot y|X does not exist it is automatically created). In addition, brushed units can be highlighted in the other following plots (only if they are already open): monitoring residual plot monitoring leverage plot maximum studentized residual $s^2$ and $R^2$ Cook distance and modified Cook distance deletion t statistics. The window style of the other figures is set equal to that which contains the monitoring residual plot. In other words, if the monitoring residual plot is docked all the other figures will be docked too. DATABRUSH IS A SCALAR: If databrush is a scalar the default selection tool is a rectangular brush and it is possible to brush only once (that is persist=''). DATABRUSH IS A CELL: If databrush is a cell, it is possible to use all optional arguments of function selectdataFS and LXS inside the curly brackets of option databrush and the following optional argument: persist = Persist is an empty value or a scalar containing the strings 'on' or 'off'. If persist = 'on' or 'off' brusing can be done as many time as the user requires. In case persist='off', every time a new brush is performed, units previously brushed are removed. In case persist='on' the unit(s) currently brushed are added to those previously brushed. However in both cases, if the user brushes a different trajectory from the one previously brushed, the previos brushed plots are stored in a figure in the background. The default value of persist is '' that is brushing is allowed only once.
bivarfit = This option adds one or more least square lines, based on SIMPLE REGRESSION of y on Xi, to the plots of y|Xi.
If bivarfit = '' is the default: no line is fitted.
If bivarfit = '1' fits a single ols line to all points of each bivariate plot in the scatter matrix y|X.
If bivarfit = '2' fits two ols lines: one to all points and another to the group of the genuine observations. The group of the potential outliers is not fitted.
If bivarfit = '0' fits one ols line to each group. This is useful for the purpose of fitting mixtures of regression lines.
If bivarfit = 'i1' or 'i2' or 'i3' etc fits an ols line to a specific group, the one with index 'i' equal to 1, 2, 3 etc. Again, useful in case of mixtures.
multivarfit = This option adds one or more least square lines, based on MULTIVARIATE REGRESSION of y on X, to the plots of y|Xi.
If multivarfit = '' is the default: no line is fitted.
If multivarfit = '1' fits a single ols line to all points of each bivariate plot in the scatter matrix y|X. The line added to the scatter plot y|Xi is avconst +Ci*Xi, where Ci is the coefficient of Xi in the multivariate regression and avconst is the effect of all the other explanatory variables different from Xi evaluated at their centroid (that is overline{y}'C)) If multivarfit = '2' exactly equal to multivarfit ='1' but this time we add the line based on the group of unselected observations.
Example: 'databrush',1
Data Types: Empty value, scalar or cell.
Output Arguments
brushedUnits —List of the
units which are inside subset in the trajectories which
have been brushed using option databrush.
brushed units. Vector. Vector
If option databrush has not been used brushedUnits will be an empty value.
References
Atkinson, A.C. and Riani, M. (2000), "Robust Diagnostic Regression Analysis", Springer Verlag, New York.
Atkinson, A.C. and Riani, M. (2002a), Tests in the fan plot for robust, diagnostic transformations in regression, "Chemometrics and Intelligent Laboratory Systems", Vol. 60, pp. 87-100.
Atkinson, A.C. Riani, M., Corbellini A. (2019), The analysis of transformations for profit-and-loss data, Journal of the Royal Statistical Society, Series C, "Applied Statistics", https://doi.org/10.1111/rssc.12389
Atkinson, A.C. Riani, M. and Corbellini A. (2021), The Box–Cox Transformation: Review and Extensions, "Statistical Science", Vol. 36, pp. 239-255, https://doi.org/10.1214/20-STS778