txmerge
txmerge performs a (hierarchical) merging of the inflated number of components found by tkmeans or tclust
Description
The function txmerge performs either a hierarchical merging of the k components found by tkmeans/TCLUST into g groups, or if g is a decimal number between 0 and 1 and DEMP is used as a distance it performs the merging phase according to such threshold.
Examples
 Example using txmerge with euclidean distances.
Example using txmerge with euclidean distances.
 Example using txmerge with euclidean distances.
Example using txmerge with euclidean distances.
close all
% Specify k cluster in v dimensions with n obs
k = 10;
v = 2;
n = 5000;
% Generate homogeneous and spherical clusters
rng(100, 'twister');
outMS = MixSim(k, v, 'sph', true, 'hom', true, 'int', [0 10], 'Display', 'off', 'BarOmega', 0.05, 'Display','off');
% Simulating data
[X, id] = simdataset(n, outMS.Pi, outMS.Mu, outMS.S);
% Plotting data
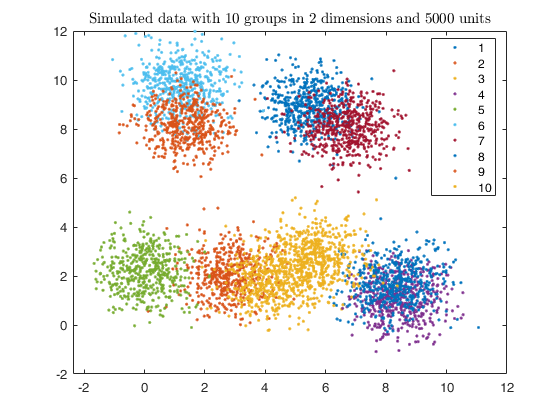
gscatter(X(:,1), X(:,2), id);
str = sprintf('Simulated data with %d groups in %d dimensions and %d units', k, v, n);
title(str,'Interpreter','Latex');
% merging algorithm based on hierarchical clustering
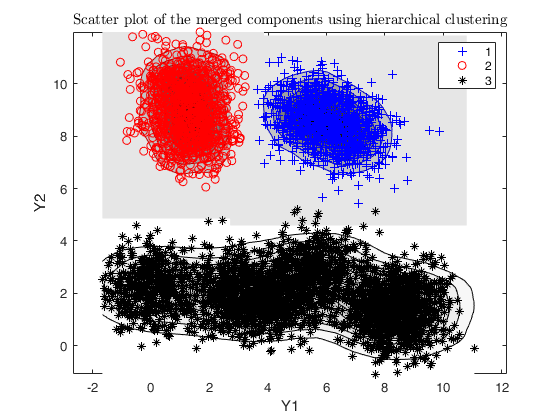
g = 3;
out = txmerge(X, k*5, g, 'dist', 1, 'plots', 'contourf');
Total estimated time to complete trimmed k means: 15.58 seconds ------------------------------ Warning: Number of subsets without convergence equal to 36.6667%
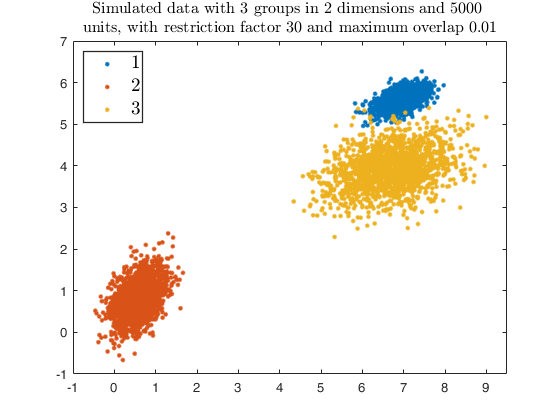
 Example using txmerge with additional arguments in the call to tkmeans.
Example using txmerge with additional arguments in the call to tkmeans.
 Example using txmerge with additional arguments in the call to tkmeans.
Example using txmerge with additional arguments in the call to tkmeans.
close all
% Specify k cluster in v dimensions with n obs
g = 3;
v = 2;
n = 5000;
% null trimming and noise level
alpha0 = 0;
% restriction factor
restr = 30;
% Maximum overlap
maxOm = 0.005;
% Generate heterogeneous and elliptical clusters
rng(500, 'twister');
outMS = MixSim(g, v, 'sph', false, 'restrfactor', restr, 'int', [0 10], ...
'Display', 'off', 'MaxOmega', maxOm, 'Display','off');
% Simulating data
[X, id] = simdataset(n, outMS.Pi, outMS.Mu, outMS.S);
% Plotting data
gg = gscatter(X(:,1), X(:,2), id);
str = sprintf('Simulated data with %d groups in %d dimensions and %d \nunits, with restriction factor %d and maximum overlap %.2f', ...
g, v, n, restr, maxOm);
title(str,'Interpreter','Latex', 'fontsize', 12);
set(findobj(gg), 'MarkerSize',10);
legend1 = legend(gca,'show');
set(legend1,'LineWidth',1,'Interpreter','latex','FontSize',14, 'Location', 'northwest')
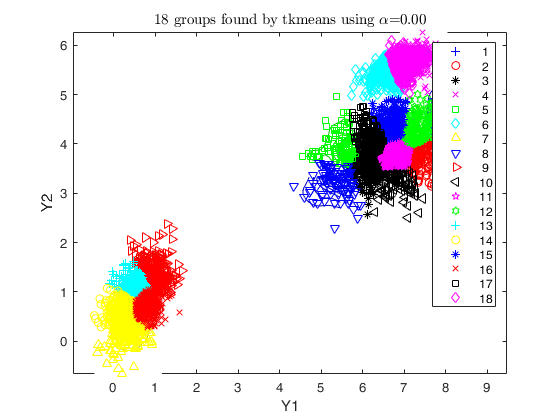
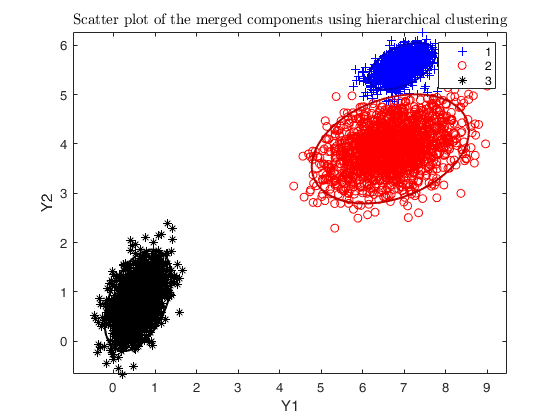
% number of components searched by tkmeans
k = g * 6;
% additional input for tkmeans
txOpt = struct;
txOpt.reftol = 0.0001;
txOpt.msg = 1;
tkmplots = struct;
tkmplots.type = 'contourf';
tkmplots.cmap = [0.3 0.2 0.4; 0.4 0.5 0.5; 0.1 0.7 0.9; 0.5 0.3 0.8; 1 1 1];
txOpt.plots = tkmplots;
txOpt.nomes = 0;
% saving tkmeans output
txOut = 1;
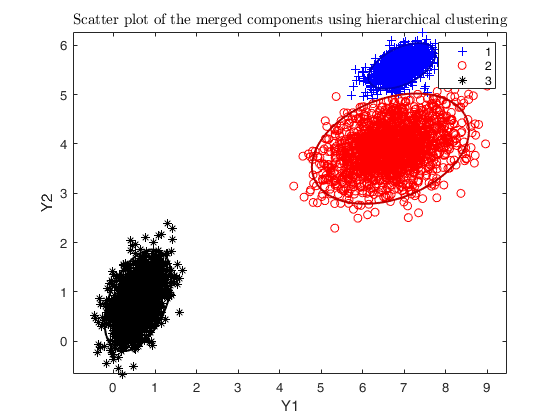
txsol = txmerge(X, k, g, 'txOpt', txOpt, 'plots', 'ellipse');
cascade;
Total estimated time to complete trimmed k means: 0.43 seconds ------------------------------ Warning: Number of subsets without convergence equal to 21.6667%
Related Examples
 Example using txmerge based on TCLUST and 'weights' linkage
close all
Specify k cluster in v dimensions with n obs
g = 3;
v = 2;
n = 5000;
null trimming and noise level
alpha0 = 0;
restriction factor
restr = 30;
Maximum overlap
maxOm = 0.
Example using txmerge based on TCLUST and 'weights' linkage
close all
Specify k cluster in v dimensions with n obs
g = 3;
v = 2;
n = 5000;
null trimming and noise level
alpha0 = 0;
restriction factor
restr = 30;
Maximum overlap
maxOm = 0.
 Example using txmerge based on TCLUST and 'weights' linkage
close all
Specify k cluster in v dimensions with n obs
g = 3;
v = 2;
n = 5000;
null trimming and noise level
alpha0 = 0;
restriction factor
restr = 30;
Maximum overlap
maxOm = 0.
Example using txmerge based on TCLUST and 'weights' linkage
close all
Specify k cluster in v dimensions with n obs
g = 3;
v = 2;
n = 5000;
null trimming and noise level
alpha0 = 0;
restriction factor
restr = 30;
Maximum overlap
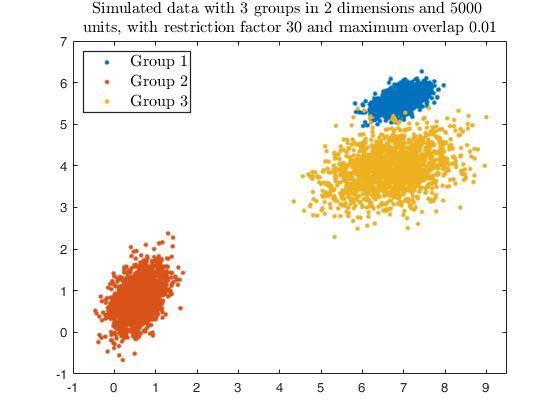
maxOm = 0.close all
% Specify k cluster in v dimensions with n obs
g = 3;
v = 2;
n = 5000;
% null trimming and noise level
alpha0 = 0;
% restriction factor
restr = 30;
% Maximum overlap
maxOm = 0.005;
% Generate heterogeneous and elliptical clusters
rng(500, 'twister');
outMS = MixSim(g, v, 'sph', false, 'restrfactor', restr, 'int', [0 10], ...
'Display', 'off', 'MaxOmega', maxOm, 'Display','off');
% Simulating data
[X, id] = simdataset(n, outMS.Pi, outMS.Mu, outMS.S);
% Plotting data
gg = gscatter(X(:,1), X(:,2), id);
str = sprintf('Simulated data with %d groups in %d dimensions and %d \nunits, with restriction factor %d and maximum overlap %.2f', ...
g, v, n, restr, maxOm);
title(str,'Interpreter','Latex', 'fontsize', 12);
set(findobj(gg), 'MarkerSize',10);
legend1 = legend(gca,'Group 1','Group 2','Group 3');
set(legend1,'LineWidth',1,'Interpreter','latex','FontSize',12, 'Location', 'northwest')
% number of components searched by tkmeans
k = g * 3;
% additional input for clusterdata (i.e. hierOpt)
linkagearg = 'weights';
txsol = txmerge(X, k, g, 'tkm', 1,'linkagearg', linkagearg, 'plots', 'ellipse');
cascade;
Total estimated time to complete trimmed k means: 0.64 seconds ------------------------------ Warning: Number of subsets without convergence equal to 37%
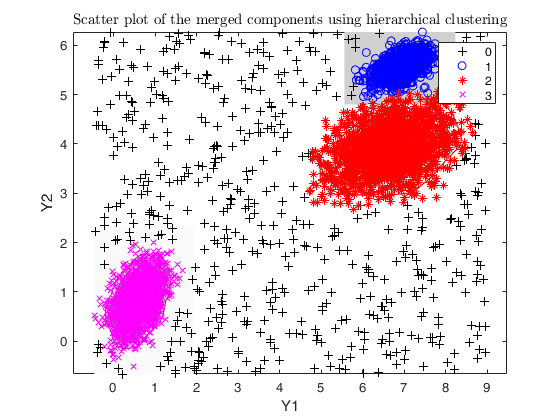
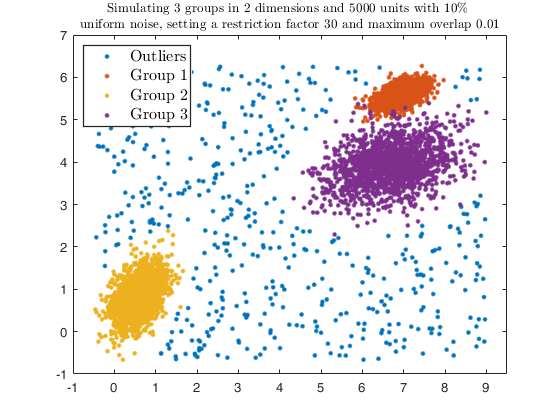
 Example using txmerge Euclidean distances or DEMP in the presence of contamination.
Example using txmerge Euclidean distances or DEMP in the presence of contamination.
 Example using txmerge Euclidean distances or DEMP in the presence of contamination.
Example using txmerge Euclidean distances or DEMP in the presence of contamination.
close all
% Specify k cluster in v dimensions with n obs
g = 3;
v = 2;
n = 5000;
% 10 percent trimming and uniform noise
alpha = 0.1;
noise = alpha*n;
% restriction factor
restr = 30;
% Maximum overlap
maxOm = 0.005;
% Generate heterogeneous and elliptical clusters
rng(500, 'twister');
outMS = MixSim(g, v, 'sph', false, 'restrfactor', restr, 'int', [0 10], ...
'Display', 'off', 'MaxOmega', maxOm, 'Display','off');
% Simulating data
[X,id] = simdataset(n, outMS.Pi, outMS.Mu, outMS.S, 'noiseunits', noise);
% Plotting data
gg = gscatter(X(:,1), X(:,2), id);
str = sprintf('Simulating %d groups in %d dimensions and %d units with %d%s \nuniform noise, setting a restriction factor %d and maximum overlap %.2f', ...
g, v, n, alpha*100, '\%', restr, maxOm);
title(str,'Interpreter','Latex', 'fontsize', 10);
set(findobj(gg), 'MarkerSize',10);
legend1 = legend(gca,'Outliers','Group 1','Group 2','Group 3');
set(legend1,'LineWidth',1,'Interpreter','latex','FontSize',12, 'Location', 'northwest')
% fixing the number of components searched by tkmeans
k = g * 6;
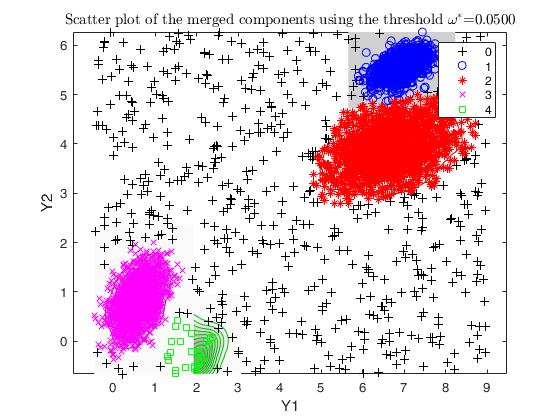
% txmerge with hierarchical merging and trimming equal to the level of noise
txsol1 = txmerge(X, k, g, 'alpha', alpha, 'plots', 'contourf');
% txmerge using a cutoff g to detect the clusters based on DEMP
g = 0.05;
txsol2 = txmerge(X, k, g, 'alpha', alpha, 'dist', 1', 'plots', 'contourf');
cascade;
Total estimated time to complete trimmed k means: 8.46 seconds ------------------------------ Warning: Number of subsets without convergence equal to 38% Total estimated time to complete trimmed k means: 6.62 seconds ------------------------------ Warning: Number of subsets without convergence equal to 36%
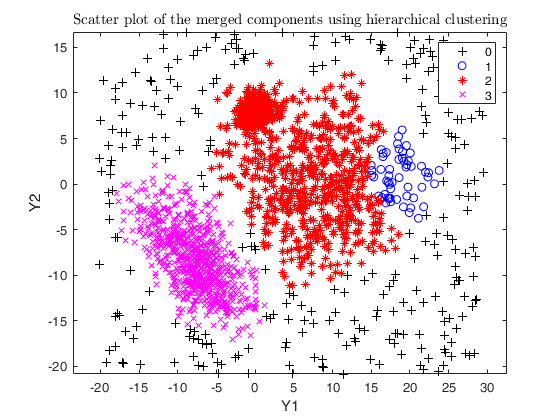
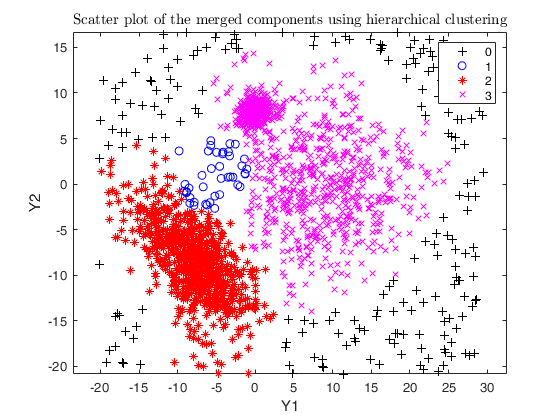
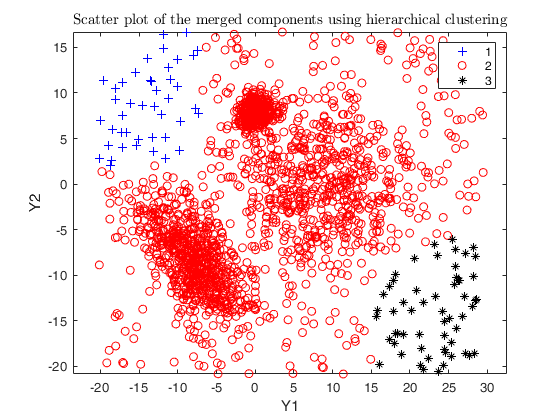
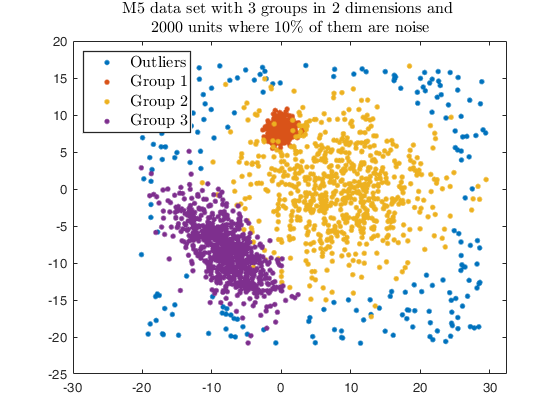

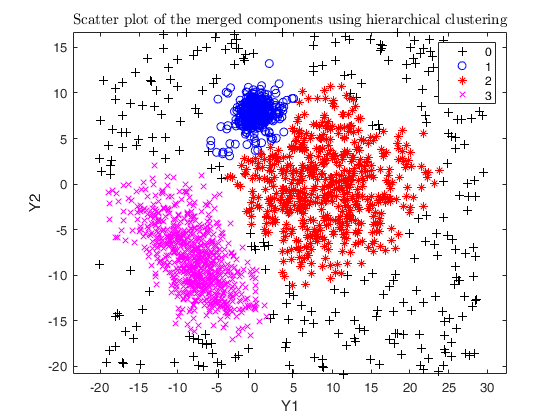
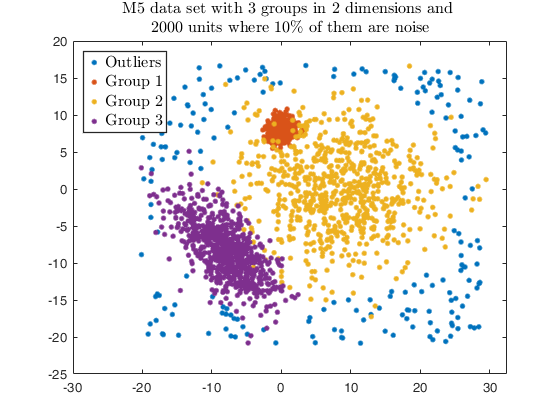
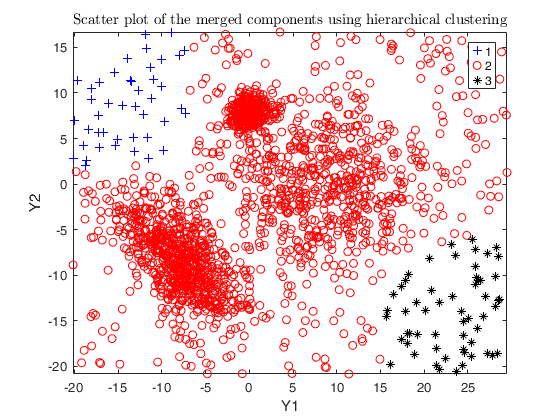
 Example using txmerge on the M5 dataset using different strategies.
Example using txmerge on the M5 dataset using different strategies.
 Example using txmerge on the M5 dataset using different strategies.
Example using txmerge on the M5 dataset using different strategies.
close all
Y = load('M5data.txt');
id = Y(:,3);
Y = Y(:, 1:2);
g = max(id);
n = length(Y);
noise = length(Y(id==0, 1));
v = 2; % dimensions
id(id==0) = -1; % changing noise label
gg = gscatter(Y(:,1), Y(:,2), id);
str = sprintf('M5 data set with %d groups in %d dimensions and \n%d units where %d%s of them are noise', g, v, n, noise/n*100, '\%');
title(str,'Interpreter','Latex', 'fontsize', 12);
set(findobj(gg), 'MarkerSize',12);
legend1 = legend(gca,'Outliers','Group 1','Group 2','Group 3');
set(legend1,'LineWidth',1,'Interpreter','latex','FontSize',12, 'Location', 'northwest')
% number of components to search
k = g*5;
% null trimming and noise level
alpha0 = 0;
% mimimum overlap cut-off value between pair of merged components
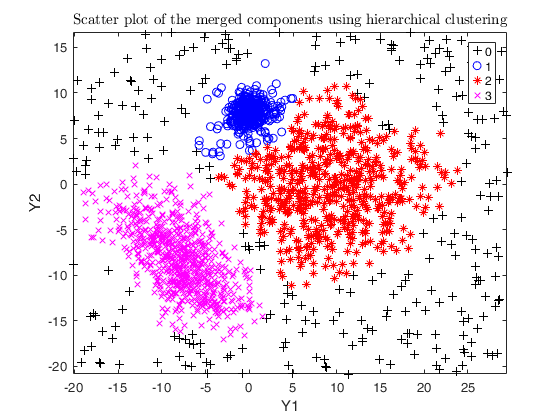
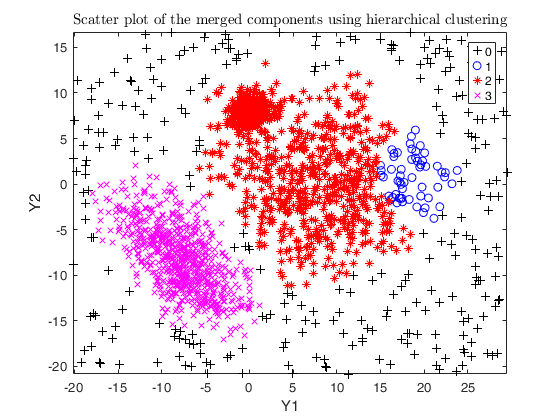
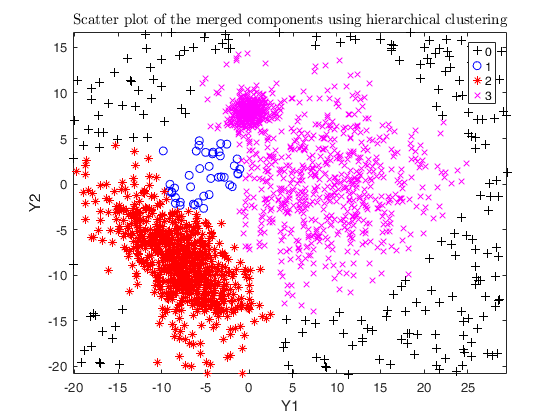
txsol1= txmerge(Y, k, g, 'alpha', alpha0, 'txOut', 1, 'plots', 1);
% setting alpha equal to noise level (usually not effective here)
alpha = noise/n;
txsol2= txmerge(Y, k, g, 'alpha', alpha, 'txOut', 1, 'plots', 1);
% setting alpha greater than the noise level
txsol3 = txmerge(Y, k, g, 'alpha', alpha+0.04, 'txOut', 1, 'plots', 1);
% using DEMP instead (usually effective)
txsol3 = txmerge(Y, k, g, 'alpha', alpha+0.04, 'txOut', 1, 'dist', 1, 'plots', 1);
cascade;
Total estimated time to complete trimmed k means: 0.16 seconds ------------------------------ Warning: Number of subsets without convergence equal to 36% Total estimated time to complete trimmed k means: 1.81 seconds ------------------------------ Warning: Number of subsets without convergence equal to 39% Total estimated time to complete trimmed k means: 1.65 seconds ------------------------------ Warning: Number of subsets without convergence equal to 38.6667% Total estimated time to complete trimmed k means: 0.12 seconds ------------------------------ Warning: Number of subsets without convergence equal to 36%




Input Arguments
Y — Input data.
Matrix.
n x v data matrix. n observations and v variables. Rows of Y represent observations, and columns represent variables. Missing values (NaN's) and infinite values (Inf's) are allowed, since observations (rows) with missing or infinite values will automatically be excluded from the computations.
Data Types: single | double
k — Number of components searched by tkmeans/TCLUST algorithms.
Integer scalar.
Data Types: single | double
g — Merging rule.
Scalar.
Number of groups retained by the hierarchical agglomeration phase, or threshold of the pairwise overlap values (i.e. omegaStar) if 0<g<1 and dist=1.
Data Types: single | double
Name-Value Pair Arguments
Specify optional comma-separated pairs of Name,Value arguments.
Name is the argument name and Value
is the corresponding value. Name must appear
inside single quotes (' ').
You can specify several name and value pair arguments in any order as
Name1,Value1,...,NameN,ValueN.
'tkm',0
, 'dist', 'squaredeuclidean'
, 'alpha', 0.05
, 'linkagearg', 'weights'
, 'auto',1
, 'plots', 1
, 'txOpt.reftol', 0.0001
, 'txOut', 1
, 'Ysave',1
tkm
—Using tkmerge or tcmerge.scalar.
Scalar. tkm=1 (default) relies on tkmeans to find an inflated number of clusters, tkm=0 is used to rely on TCLUST instead.
Example: 'tkm',0
Data Types: double
dist
—Distance between clusters.scalar, char.
Its value indicates the merging rule for the initial number of (inflated) clusters. If dist=0 the distance between centroids is Euclidean (default), if dist=1 directly estimated misclassification probabilities (DEMP) are used, else use a character according to MATLAB pdist function.
Example: 'dist', 'squaredeuclidean'
Data Types: single | double | string
alpha
—Global trimming level.scalar.
alpha is a scalar between 0 and 0.5. If alpha=0 (default) tkmeans reduces to kmeans and TCLUST reduces to MCLUST.
Example: 'alpha', 0.05
Data Types: single | double
linkagearg
—Linkage used for hierarchical agglomeration.single linkage is the default, see the MATLAB linkage function for other options.
Example: 'linkagearg', 'weights'
Data Types: character
auto
—Automatic trimming level detection.scalar.
It is set to 1 to overwrite the prespecified alpha parameter, or it is equal to 0 to use alpha as trimming level (default).
Example: 'auto',1
Data Types: double
plots
—Plot on the screen.scalar, char, | struct.
- If plots=0 (default) no plot is produced.
- If plots=1, the components merged are shown using the spmplot function. In particular: * for v=1, an histogram of the univariate data.
* for v=2, a bivariate scatterplot.
* for v>2, a scatterplot matrix.
When v>=2 plots offers the following additional features (for v=1 the behaviour is forced to be as for plots=1).
If plots is a char it may contain the words 'contourf', 'contour' 'ellipse' and 'boxplotb'. For the documentation of these option see below the case when plots is a structure If plots is a structure it may contain the following fields:
| Value | Description |
|---|---|
type |
Type of plot to add in the background or to superimpose. It can be: 'contourf', 'contour', 'ellipse' or 'boxplotb', specifying respectively to add filled contour (default when overlay=1), contour, ellipses or a bivariate boxplot (see function boxplotb.m). - plots.type='contourf' adds in the background of the bivariate scatterplots a filled contour plot. The colormap of the filled contour is based on grey levels as default. - plots.type='contour' adds in the background of the bivariate scatterplots a contour plot. The colormap of the contour is based on grey levels as default. - plots.type='ellipse' superimposes confidence ellipses to each group in the bivariate scatterplots. The size of the ellipse is chi2inv(0.95,2), i.e. the confidence level used by default is 95%. - plots.type='boxplotb' superimposes on the bivariate scatterplots the bivariate boxplots for each group, using the boxplotb function. |
cmap |
colors to use. Three-column matrix of values in the range [0,1] where each row is an RGB triplet that defines one color. Check the colormap function for additional information. REMARK - The labels<=0 are automatically excluded from the overlaying phase, considering them as outliers. |
Example: 'plots', 1
Data Types: double | struct
txOpt
—tkmeans/TCLUST optional arguments.structure.
Empty structure (default) or structure containing optional input arguments for tkmeans or TCLUST. See tkmeans and tclust functions.
Example: 'txOpt.reftol', 0.0001
Data Types: struct
txOut
—Saving tkmeans/TCLUST output structure.scalar.
It is set to 1 to save the output structure of tkmeans/TCLUUST into the output structure of txmerge. Default is 0, i.e. no saving.
Example: 'txOut', 1
Data Types: single | double
Ysave
—Saving Y.scalar.
Scalar that is set to 1 to request that the input matrix Y is saved into the output structure out.
Default is 0, i.e. no saving is done.
Example: 'Ysave',1
Data Types: double
Output Arguments
out — description
Structure
Structure which contains the following fields
| Value | Description |
|---|---|
PairOver |
Distance matrix among the k components found by tkmeans/TCLUST. |
mergID |
Label for each unit. It is a vector with n elements which assigns each unit to one of the groups obtained according to the merging algorithm applied. REMARK - out.mergID=0 denotes trimmed units. |
txOut |
Output from tkmeans function. This structure is present only if option txOut is set to 1. |
Y |
Original data matrix Y. This field is present only if option Ysave is set to 1. |
References
Insolia, L., Perrotta, D. (2023), Tk-Merge: Computationally Efficient Robust Clustering Under General Assumptions. Advances in Intelligent Systems and Computing, vol 1433. Springer, Cham.
See Also
dempk
|
tkmeans
|
clusterdata
|
tclusteda
|
overlapmap