carbikeplotGPCM
carbikeplotGPCM produces the carbike plot to find best relevant clustering solutions
Description
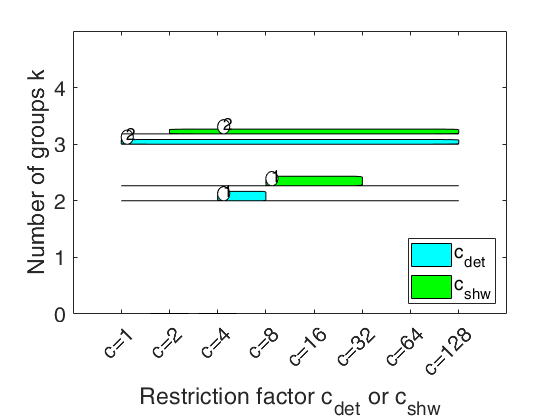
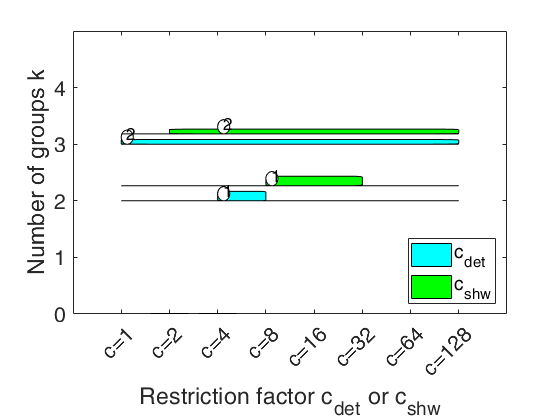
carbikeplotGPCM takes as input the output of function tclustICsolGPCM (that is a structure containing the best relevant solutions) and produces the car-bike plot. This plot provides a concise summary of the best relevant solutions. This plot shows on the horizontal axis the value of and c_{shw} restriction factors and on the vertical axis the value of k. For each solution we draw two rectangle (associated with c_{det} and c_{shw}) which are respectively referred to interval of values for which the solution is best and stable and a horizontal line which departs from the rectangle for the values of c_{det} (c_{shw}) in which the solution is only stable.
Finally, for the best value of c_{det} (c_{shw}) associated to the solution, we show a circle with a number indicating the ranked solution among those which are not spurious. This plot has been baptized ``car-bike'', because the first best solutions (in general 2 or 3) are generally best and stable for a large number of values of c and therefore will have large rectangles. In addition, these solutions are likely to be stable for additional values of c_det (c_{shw}) and therefore are likely to have horizontal lines departing from the rectangles (from here the name ``cars''). Finally, local minor solutions (which are associated with particular values of c_{det} (c_{shw}) and k) do not generally present rectangles or lines and are shown with circles (from here the name ``bikes'')
car-bike plot for the geyser data.h
=carbikeplotGPCM(RelSol,
Name, Value)
Examples
Input Arguments
Output Arguments
References
Cerioli, A. Garcia-Escudero, L.A., Mayo-Iscar, A. and Riani, M. (2017), Finding the Number of Groups in Model-Based Clustering via Constrained Likelihoods, "Journal of Computational and Graphical Statistics", pp. 404-416, https://doi.org/10.1080/10618600.2017.1390469
 Simulated data with 3 components.
Simulated data with 3 components.